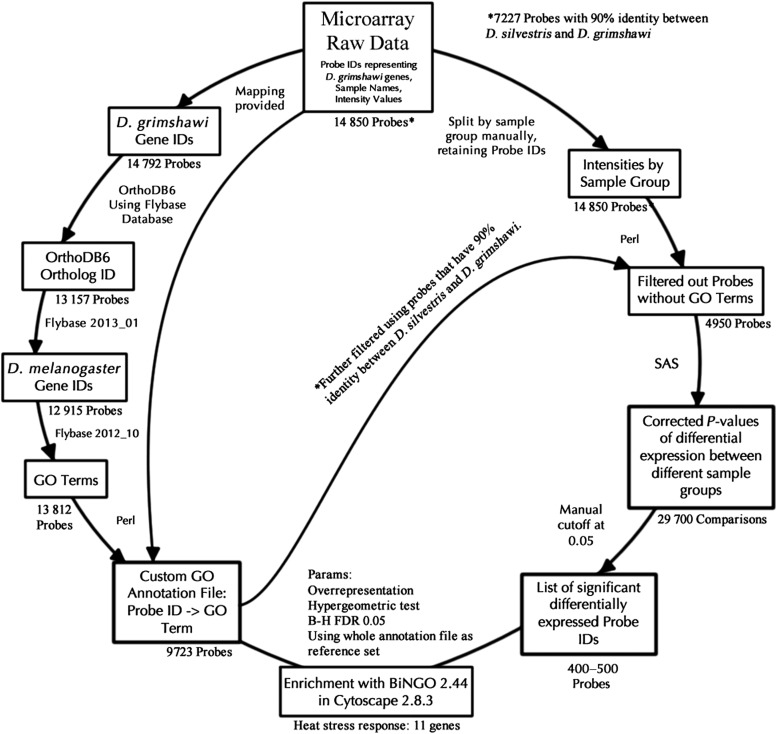
Figure 1:
Bioinformatics workflow used to detect enriched Gene Ontology (GO) terms between 16 and 25°C treatments of Hawaiian picture-wing Drosophila. Drosophila grimshawi GO annotations were associated with the microarray probes using OrthoDB6. These values were then associated with array intensities, and a hierarchical linear model was used to generate the list of significant differentially expressed probes. These results were used in BiNGO to identify enriched GO terms. * The analysis was done with the original 14 792 probes and with the 7227 probes that had 90% identity between D. silvestris and D. grimshawi.