Fig. 1.
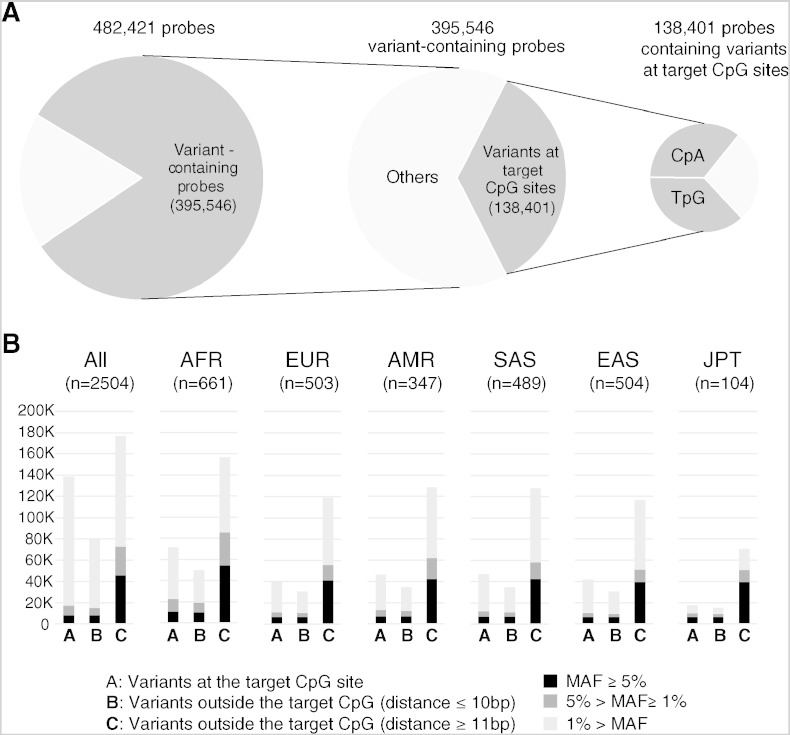
Numbers of HM450 probes whose genomic interval overlaps with nucleotide variants reported in the Phase 3 dataset of the 1000 Genomes Project. A. Out of all 482,421 probes that target a CpG site, 395,546 probes contained one or more reported variant(s) in their genomic intervals. Among them, 138,401 probes (35.0%) overlapped with nucleotide variant(s) at their target CpG sites. Out of the 138,401 sites, alternate dinucleotides of 100,523 sites (72.6%) were either TpG or CpA. B. Numbers of probes containing nucleotide variants detected among Japanese (JPT), African (AFR), European (EUR), American (AMR), South Asian (SAS), and East Asian (EAS) subjects, and among all subjects (n = 2500). In each panel, variant-containing probes were classified in three categories (A, B, and C) depending on the distance to the C (MAPINFO [4]) of the target CpG site. In each category, probes were further divided into three sub-categories depending on the minor allele frequency of the overlapping variant(s) and shown as stacked column charts (black, MAF ≥ 5%; dark gray, 5% > MAF ≥ 1%; light gray, 1% > MAF).