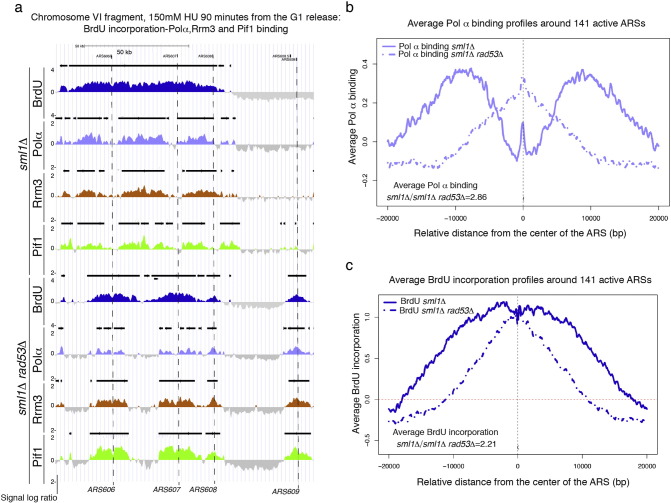
Fig. 1.
) a) BrdU incorporation (dark blue) and Polα-Flag (light blue), Rrm3-13Myc (brown) and Pif1-Flag (green) binding profiles were determined, respectively, by ssDNA-BrdU IP on chip and ChIP on chip in the strains CY12488, CY13284, CY12470, CY13074, CY12493, CY13282, CY12422 and CY13073 released from G1 into 150 mM of HU for 90 min [9]. The y-axis shows the enrichment signals expressed as ratio log2 IP/SUP of loci significantly enriched in the IP fractions. The horizontal black bars above the picks indicate statistically significative BrdU or binding clusters. X-axis represents chromosomal coordinates. Early DNA replication origins (ARS606, ARS607) and the dormant origins (ARS608 and ARS609) are marked by dashed black lines. A black scale bar on the chromosome map indicates 50 kbps. b) Average binding profiles of DNA polymerase α in sml1Δ and sml1Δ rad53Δ cells (from the experiment shown in panel a), in a window of 40 kbps centered on each of the 141 active ARSs are shown. The ratio of average Pol α binding signals in sml1Δ versus sml1Δ rad53Δ cells is 2.86. c) Average BrdU incorporation profiles in sml1Δ and sml1Δ rad53Δ cells in a window of 40 kbps centered on 141 ARSs have been determined in the experiment shown in panel a. The ratio of BrdU incorporation signals in sml1Δ versus sml1Δ rad53Δ cells around 141 ARSs is 2.21.