Abstract
Recently, for copy number variation (CNV) analysis, bovine researchers have focused mainly on the use of genome-wide SNP genotyping arrays. One of the highest densities commercially available SNPchips for cattle is the Affymetrix axiom genome-wide Bos 1, which assays 648,315 informative SNPs across the whole bovine genome. Here, we describe the microarray data, quality controls and validation implemented in a study published in Genetics and Molecular Research Journal in 2015 [1]. The microarray raw data has been deposited into Gene Expression Omnibus under accession #GSE54813.
Keywords: Holstein cattle, Copy number variation, SNP, Axiom genome-wide Bos 1 array, Bioinformatics, PennCNV
| Organism/cell line/tissue | Bos taurus |
| Sex | Female |
| Sequencer or array type | Affymetrix axiom genome-wide Bos 1 array |
| Data format | Raw data |
| Experimental factors | Any pretreatment of samples |
| Experimental features | DNA extraction from blood samples cows, genotyped with high density arrays, copy number variations detection and validation. |
| Consent | Not applicable. |
| Sample source location | Veterinary Science Research Institute of the Autonomous University of Baja California, Mexicali, Mexico. |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Characteristics of the samples
The blood samples were collected by venipuncture of the coccygeal vein, from 12 Holstein dairy cows, registered in the Mexican Holstein Association. All were born after artificial insemination, and were between their first and fourth lactation; they were all clinically healthy and free of brucellosis and tuberculosis. All were selected such that they were not related in the last three generations.
2.2. DNA extraction and genotyping
DNA extraction and purification were performed using a QIAGEN kit. All DNA samples were analyzed by spectroscopy and agarose gel electrophoresis, and were genotyped with the axiom genome-wide Bos 1 array with an average call rate for each individual sample of 99.7%. The raw data of the SNPchip were submitted to the Gene Expression Omnibus under the accession number GSE54813.
2.3. Microarray data processing
We extracted signal intensity (SI) and B allele Frequency (BAF) values from CEL files (raw data) for each SNP. Values were generated by the Affymetrix Power Tools (APT) software, which implements a set of cross-platform command line algorithms for analyzing and working with Affymetrix arrays. APT documentation can be obtained from (http://www.affymetrix.com/estore/ partners_programs/programs/developer/tools/powertools.affx). We also used the guideline defined in PennCNV-Affy Protocol for CNV detection in Affymetrix SNP arrays (http://penncnv.openbioinformatics.org/en/latest.user-guide/affy/).
2.4. Normalization and quality control
The intensity values from the two alleles are referred as the A and B alleles. These alleles are summarized signal intensity values obtained from “AxiomGT1.summary.txt” file, produced by the APT software. Finally, the two values of signal intensity for each SNP were normalized by expressing them as Log2 ratio (LRR), using a Perl script which implements the following procedure: first, a reference is developed for each marker considering the formula T = A + B, where A and B are the values of the signal intensity of each allele. For each SNP, a reference to the value M = median is set (T_sample1, T_Sample2, …, T_sampleN). The second step is to estimate the intensity for each individual sample with the formula log2 (T/M) ratio, from which we get the normalized signal intensity for each SNP and each sample SNP [2].
We applied some quality control filters to the data, we eliminated all SNPs with genotyping errors (no call), based on the “AxiomGT1.calls” file, which contain genotype calls (− 1 = NN, AA = 0, AB = 1, BB = 2). We also filtered all non-somatic SNPs. Our final working dataset was of 601,894 SNPs.
2.5. Genome-wide identification of CNVs
We used two algorithms: PennCNV [3] and QuantiSNP [4], for CNV detection. The PennCNV algorithm requires as input LRR and BAF values for each marker, and the distance between each SNP. PennCNV was executed using default values for the 29 autosomal chromosomes, and genomic waves were adjusted using the argument called GC model. The GC model file for this study was generated by a Perl script, which computes the GC content within 1 Mb around each marker (500 kb each side). QuantiSNP was executed with the options -isaffy and -levels enabled since we used an Affymetrix array. In the same way -gcdir option was enabled to perform the correction of the LRR, in markers affected by genomic waves [5] (Fig. 1).
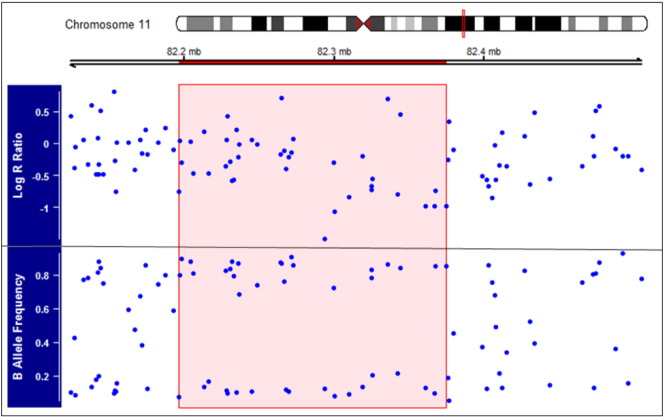
Fig. 1.
Log R ratio (LRR) and B allele frequency (BAF) plot of one copy number variation region (CNVR). Inside the selected area, low values of LRR (less than − 1) and no values in the 0.5 cluster indicate a single copy deletion in a region of chromosome 11.
For declaring a putative CNV, we considered at least three adjacent SNPs indicating a loss or gain, with a total length greater or equal to 1 kb, detected simultaneously by the two algorithms in the same animal, either in the same position or overlapping. Finally, CNV regions (CNVRs) were defined based on the criteria used in a study by Redon et al. [6].
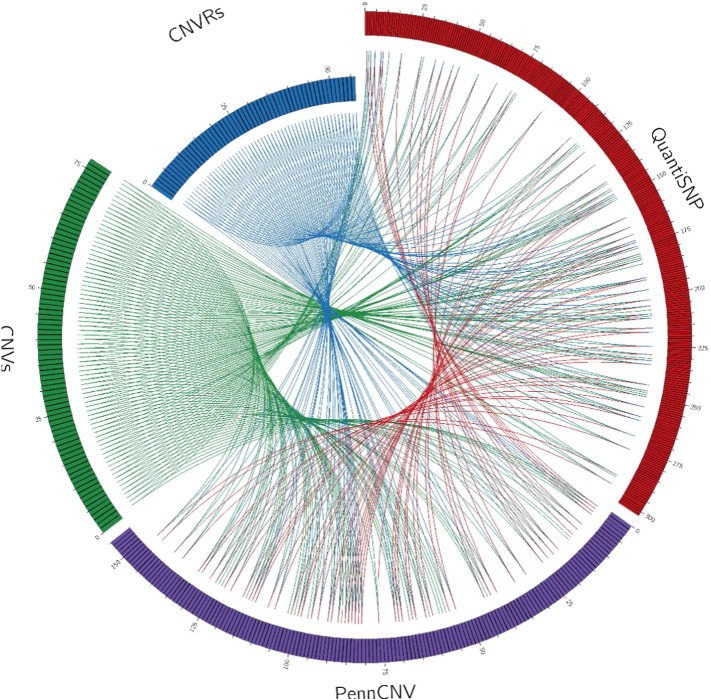
PennCNV detected 155 CNVs, while QuantiSNP detected 302. The algorithms coincided for 77 putative CNVs, detected in the same position and the same sample (Fig. 1). Initially, we termed these variants as putative CNVs. We inspected the 77 CNVs for overlaps and defined 56 CNVRs. (Fig. 2).
Fig. 2.
The circle shows the coincidences of CNVs and CNVRs between the two algorithms. Links on blue and red represents the 56 CNVRs where PennCNV and QuantiSNP coincided. Links on green represents the 77 CNVs in which both algorithms coincided.
3. Basic analysis
3.1. Functional analysis of genes
To identify gene contents and to obtain a description of each gene affected within the regions covered by CNVRs, we used the BioMart database (http://www.biomart.org) and the RefGen database (http://refgene.com). We found 103 genes, of which 96 encoded proteins, two were pseudogenes, three were snRNAs, and two were miRNAs. In order to analyze functional enrichment in the CNVRs, we searched the Gene Ontology (GO) database [7] and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [8]. Both analyses were carried out using the bioinformatic tool DAVID [9]. The GO analysis showed common gene terms among mammals. KEGG pathway analysis showed that the genes were mainly represented in the pathway of olfactory transduction.
3.2. CNV validation by real-time PCR (qPCR)
For each target CNVR, two pairs of primers were designed considering the limits of each CNVR. PCR primers were designed using the NCBI Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast). The PCR amplification program was 5 min at 95 °C, followed by 40 cycles at 95 °C for 10 s and 60 °C for 10 s. We used the Basic Transcription Factor (BTF3) as a control gene for comparing the number of copies in each CNVR [1].
We use the method of comparative cycle threshold (2− ΔΔCt) to quantify the number of changes of the copies by comparing the ΔCt value, from the samples with CNV to a ΔCt of a calibrator without CNV [10]. The average Ct value of three replicates for each sample was calculated, normalized, and compared against the control gene, with the assumption of the existence of two copies of the DNA segment in the control region.
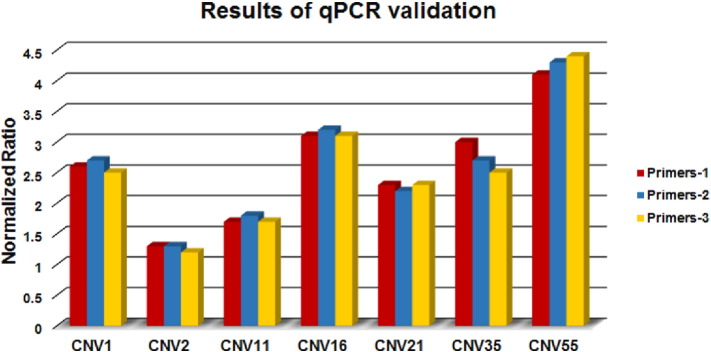
For each CNVR to be validated, the value of 2 × 2− ΔΔCt was calculated for each individual. The obtained value was used to decide if a CNVR was normal (without CNVR, if the value was about two), or a gain (if the value was about three or above), or a deletion (if the value was near zero or one) [11] (Fig. 3).
Fig. 3.
The normalized ratio in two (normal state), assumes the existence of two copies of the DNA segment. Values around one indicate a single copy loss; values around three indicate a three copy gain, and around four indicate a four copies gain.
4. Discussion
We describe here, in the best of our knowledge, the first publically available high-density SNP genotypes dataset from bovine genome. This dataset is composed of raw data from 12 Holstein cows. 56 CNVRs genome-wide were identified in the analysis. In addition, five of the putative CNVRs were validated by qPCR. Finally, we showed that SNP data from Affymetrix axiom genome-wide Bos 1 array, allows achieving great accuracy in the identification of CNVRs and their candidate genes.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We are grateful to the Council for Science and Technology of Mexico (CONACYT) for supporting a scholarship for postdoctoral studies for Ricardo Salomon-Torres, number scholarship 362690.
References
- 1.Salomon-Torres R., Gonzalez-Vizcarra V.M., Medina-Basulto G.E., Montano-Gomez M.F., Mahadevan P., Yaurima-Basaldua V.H. Genome-wide identification of copy number variations in Holstein cattle from Baja California, Mexico, using high-density SNP genotyping arrays. Genet. Mol. Res. 2015;14:11848. doi: 10.4238/2015.October.2.18. [DOI] [PubMed] [Google Scholar]
- 2.Rincon G., Weber K.L., Eenennaam A.L., Golden B.L., Medrano J.F. Hot topic: performance of bovine high-density genotyping platforms in Holsteins and Jerseys. J. Dairy Sci. 2011;94:6116. doi: 10.3168/jds.2011-4764. [DOI] [PubMed] [Google Scholar]
- 3.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colella S., Yau C., Taylor J.M., Mirza G., Butler H., Clouston P. QuantiSNP: an objective Bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diskin S.J., Li M., Hou C., Yang S., Glessner J., Hakonarson H. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D. Global variation in copy number in the human genome. Nature. 2006;444:444. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Jiang L., Jiang J., Wang J., Ding X., Liu J., Zhang Q. Genome-wide identification of copy number variations in Chinese Holstein. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048732. [DOI] [PMC free article] [PubMed] [Google Scholar]