Figure 1. Similar to Nupr1, Nurp1L displays properties congruent with their role in gene expression regulation.
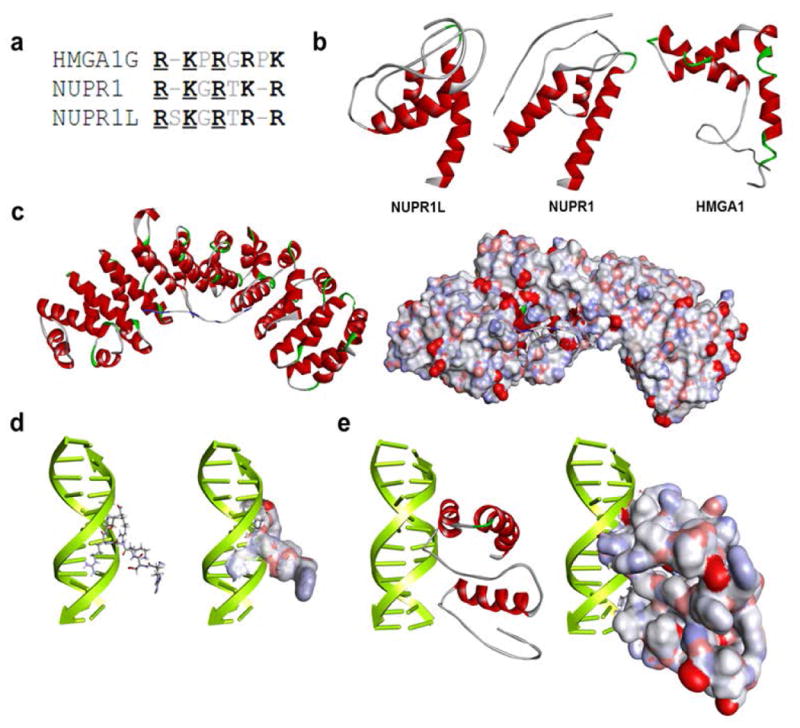
(a) Comparison between DNA-binding domains of NUPR1L and NUPR1 with the AT-hook of HMGA1.(b) NUPR1L and its closest human homolog NUPR1 share their ability to adopt a helix-loop-helix fold that is characteristic of HMGA1. Ribbon representation of their models with helical regions denoted in red and loops in white. (c) Homology-based models derived from the PDB complex 1EJY. The models show that a C-terminal peptide sequence of NUPR1L (KRVAQKLLRGQRKRR) kinds to alpha-importin which mediates the nuclear localization of most eukaryotic proteins. The ribbon model shows the C-terminal peptide from NUPR1L with blue coloring n basic regions that form the most bonds with the main chain of alpha-importin. A space-filling model displays the cavity within alpha-importin which easily accommodates the C-terminal peptide of NUPR1L. (d) Two different molecular modeling approaches were used to gain insight into the DNA-binding properties of NUPR1L. Both a homology-based approach and a DP-DOCK-based docking approach and scoring method reveal that NUPR1L binds to the minor groove of DNA using a DNA binding sequence (GRSKGRTRR) that is highly similar to the AT-hook containing transcriptional activator HMGA1. HMGA1-based homology models in a ribbon and space-filling representation are shown. (e) DP-DOCK-based model of full-length NUPRL are shown.
