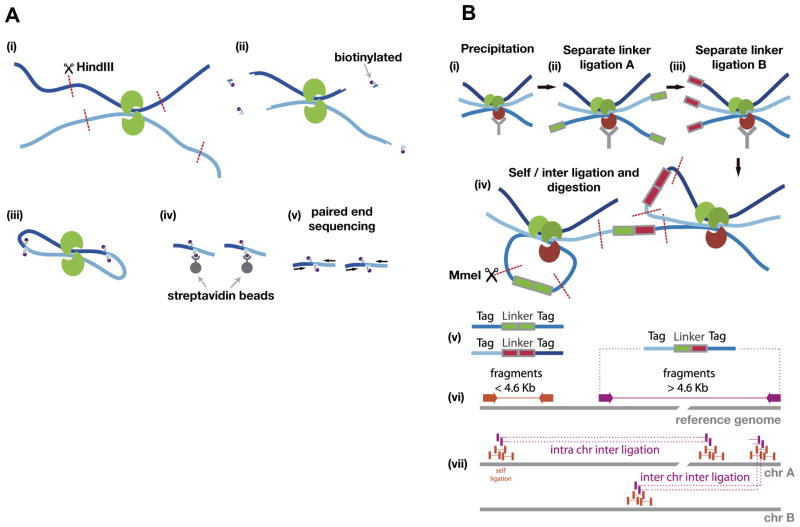
Fig. 3.
Overview of Hi-C and ChIA-PET. (A) A schematic shows the Hi-C protocol. (i) Formaldehyde cross-linking of the cells (Proteins in green, Chromatin dark blue and light blue), followed by digestion (HindIII) of the chromatin. (ii) The restriction site is used to attach biotinylated nucleotides (purple). (iii) Ligation of the open ends. (iv) Streptavidin beads are used to isolate the biotinylated molecules, followed by (v) paired-end sequencing. (B) Schematic of ChIA-PET analysis: The chromatin is prepared by formaldehyde cross-linking, fragmentation (not shown) and (i) precipitation, followed by (ii), (iii) separate linker ligation A and B. Then, the separate probes are mixed to allow for proximity (inter) and self-ligation, which is followed by (iv) MmeI restriction enzyme digestion. After sequencing, the resulting tag-linker products (v) are mapped to the genome (vi). The linker can be used to categorize between self and inter ligation, which allows for clustering of self-ligation and long-range chromatin interactions (vii).