Extended Data Figure 4. Isolation of PS fractions from Arabidopsis seedlings and LIMYB-mediated inhibition of protein synthesis.
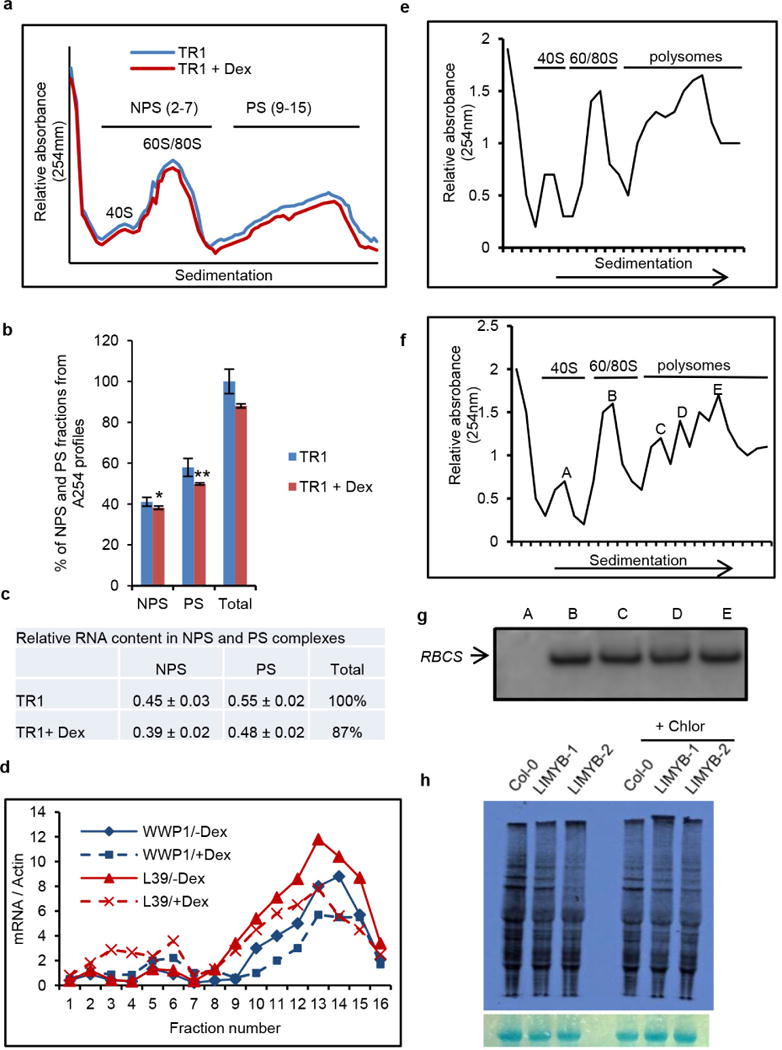
a, Ultraviolet absorbance profiles of the sucrose gradient used for RNA fractionation of dexamethasone (Dex)-inducible T474D transgenic lines. Sixteen fractions of 310 ml were collected. NPS RNA includes complexes ≤ 80S that fractionated in the top half of the gradient (fractions 2–7) and PS (polysome) represents complexes fractionated in the bottom of the gradient (fractions 9–15). b, Distribution (%) of NPS and PS fractions on polysome density gradients. The percentage of NPS or PS was calculated by integrating the areas under the corresponding peaks in the A254nm profile delimited by a gradient baseline absorbance (buffer density). Values are the average ± standard deviation (s.d.) of three biological replicates. NPS and PS fractions from T474D + Dex samples were significantly different from the corresponding fractions of the T474D samples by the t-test (greater). NPS, *P value = 0.03725; PS, **P value = 0.009137 (t-test; greater). c, Relative RNA content in NPS and PS complexes. RNA was precipitated from NPS and PS density regions of sucrose gradients (as in a) and quantified. Relative NPS RNA and PS RNA contents from T474D and T474D + Dex samples were calculated in relation to the total NPS + PS content from T474D. Values for the relative NPS and PS RNA content are the average ± s.d. of three biological replicates and they were significantly different between the samples (P < 0.01, t-test). d, Distribution of specific mRNAs in the PS gradient fractions from extracts prepared from T474D seedlings treated (or not) with dexamethasone. The RNA on each fraction was reverse transcribed and aliquots amplified with specific primers for the indicated genes by qPCR. e, Ultraviolet absorbance profiles of the sucrose gradient used for the RNA fractionation of Col-0 seedlings. PSs from 15-day-old Col-0 seedlings were fractionated on a sucrose gradient, and the fractions were manually collected. f, Ultraviolet absorbance profiles of the sucrose gradient used for RNA fractionation from T474D seedlings. PSs from 15-day-old T474D-overexpressing seedlings were fractionated on a sucrose gradient, and the fractions were manually collected. g, Levels of the small subunit of rubisco (RBCS) mRNA per fraction. The levels of mRNA of RBCS were examined by northern blotting. This control was used to ensure the quality and distribution of a specific mRNA. h, Overexpression of LIMYB suppresses cytosolic translation. In vivo labelling of leaf proteins with [35S]Met was performed in Col-0 and LIMYB-1 transgenic seedlings in the presence and absence of chloramphenicol treatment. The total protein extracts were fractionated by SDS–PAGE, and the radioactive bands were quantified by densitometric analysis of the images obtained by autoradiography. The labelling percentage was normalized to the leaf chlorophyll content, and protein loading is shown by Coomassie staining of the radioactive gel.
