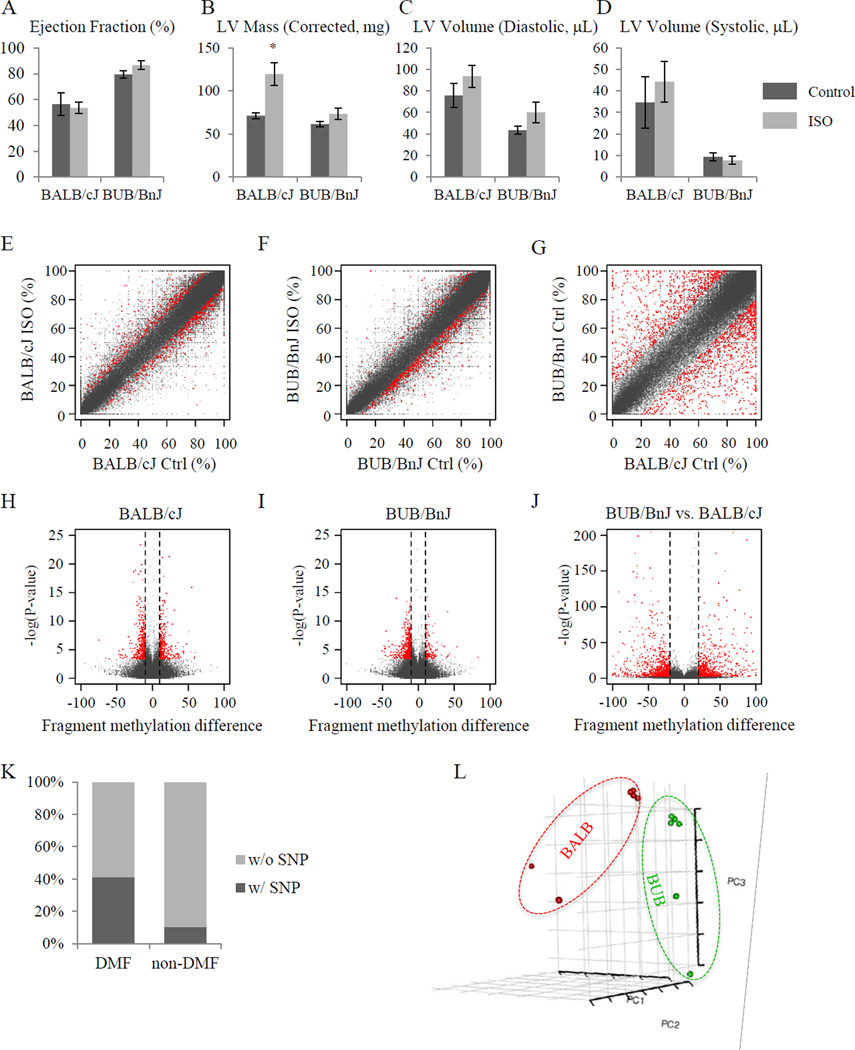
Figure 2. ISO-treatment induces differential DNA methylation.
(A–D) Isoproterenol induced cardiac pathology in mice of BALB/cJ and BUB/BnJ strains indicating strain-specific responses to beta-adrenergic receptor agonist treatment. The heart function as indicated by ejection fraction (A) and size of heart as indicated by left ventricular mass (B), diastolic left ventricular volume (C) and systolic left ventricular volume (D), were measured using M-mode echocardiography. BALB/cJ mice showed a dramatic increase of LV mass and volume after three-week ISO treatment, while BUB/BnJ mice showed only a slight increase of ejection fraction. (n=5–13/group for panels A–D. *P<0.05 vs. control. Vertical bars represent Standard Error) (E–G) Scatter plots showing differential DNA methylation per fragment, i.e. MspI digestion product with length of 50bp to 300bp. Axes indicate DNA methylation level in percentage. N=3/group for all methylation studies. (E) ISO-induced differential DNA methylation in BALB/cJ strain. (F) ISO-induced differential DNA methylation in BUB/BnJ strain (see Online Figure I for individual CpG data). (G) Basal difference between BALB/cJ and BUB/BnJ. (H-J) Volcano plot showing ISO-induced differentially methylated fragments in BALB/cJ (H) and BUB/BnJ (I) strains, as well as basal differences between BALB/cJ and BUB/BnJ (J). x-axis, difference of DNA methylation level between control and ISO treated mice. y-axis, −log10 of p-value. Red, differentially methylated DNA fragments with FDR < 5%. For a CpG methylation event to be considered different in panels H & I (red data points), the site must exhibit a >10% (>20% for basal comparison, panel J) difference with FDR < 0.05. (K) Stacked column plot showing the percentage of DNA fragments harboring at least one genomic variance (SNP) between BALB/cJ and BUB/BnJ. DNA fragments from the whole genome (left column) and with differential methylation levels between two strains at basal level (right column) are shown. P-value was calculated using Pearson’s Chi-squared test with Yates’ continuity correction, and is smaller than 2.2×10−16. (L) Principal component analysis of fragment CpG methylation is shown with the first three principal components, demonstrating a strain-specific DNA methylome profile.