Fig. 2. The peroxisomal protease LON2 inhibits pexophagy in Arabidopsis.
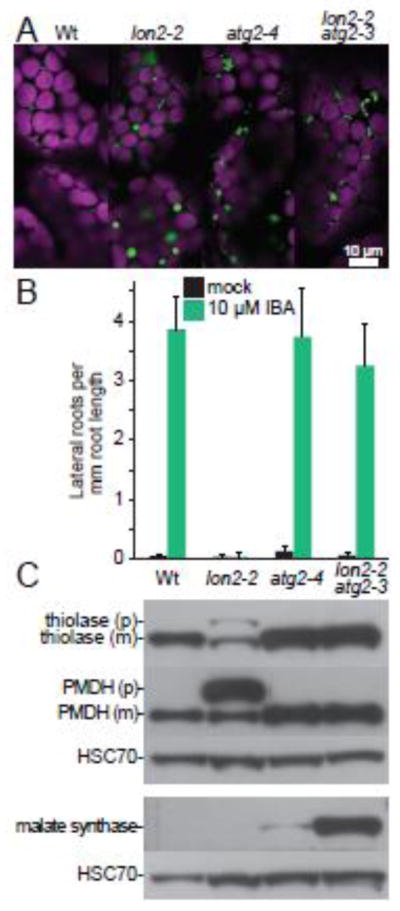
A) Preventing autophagy suppresses protein import defects and the large puncta phenotype of lon2-2. Cotyledon mesophyll cells in 8-d-old seedlings of the indicated genotypes expressing 35S:PTS2-GFP [81] were imaged for fluorescence using confocal microscopy. PTS2-GFP bears an N-terminal PTS2 that localizes GFP (green) to peroxisomes in wild-type Columbia-0 (Wt), atg2-4 and lon2-2 atg2-3. PTS2-GFP is partially cytosolic (diffuse fluorescence) in lon2-2. Peroxisomes are enlarged in lon2-2 and clustered in atg2-4 and lon2-2 atg2-3. Chlorophyll autofluorescence (magenta) marks chloroplasts. Scale bar = 10 μm.
B) Preventing autophagy suppresses the IBA resistance of lon2-2. Wild-type Columbia-0 (Wt), lon2-2 [38], atg2-4 [28], and lon2-2 atg2-3 [28] seeds were plated on medium containing 0.5% sucrose (mock) and grown for 4 days. Half of the seedlings were then transferred to medium supplemented with 0.5% sucrose and 10 μM IBA, and seedlings were grown for an additional 4 days before measuring lateral root formation. Error bars represent standard deviation of the means (n ≥ 10).
C) Preventing autophagy suppresses the PTS2-processing defect of lon2-2, and obsolete matrix proteins are stabilized in lon2-2 atg2-3 double mutants. Extracts from 6-day-old light-grown seedlings of the indicated genotypes were processed for immunoblotting in duplicate, and membranes were serially probed with antibodies recognizing the peroxisome matrix proteins thiolase [13] or malate dehydrogenase [PMDH; 82], shown in the upper panel, or malate synthase [MLS; 83], shown in the lower panel. Thiolase and PMDH are PTS2 proteins synthesized as precursors (p) in the cytosol and cleaved to mature forms (m) lacking the PTS2 region in the peroxisome. Protein loading was monitored by probing with an antibody recognizing HSC70 (SPA-817; StressGen Bioreagents).
