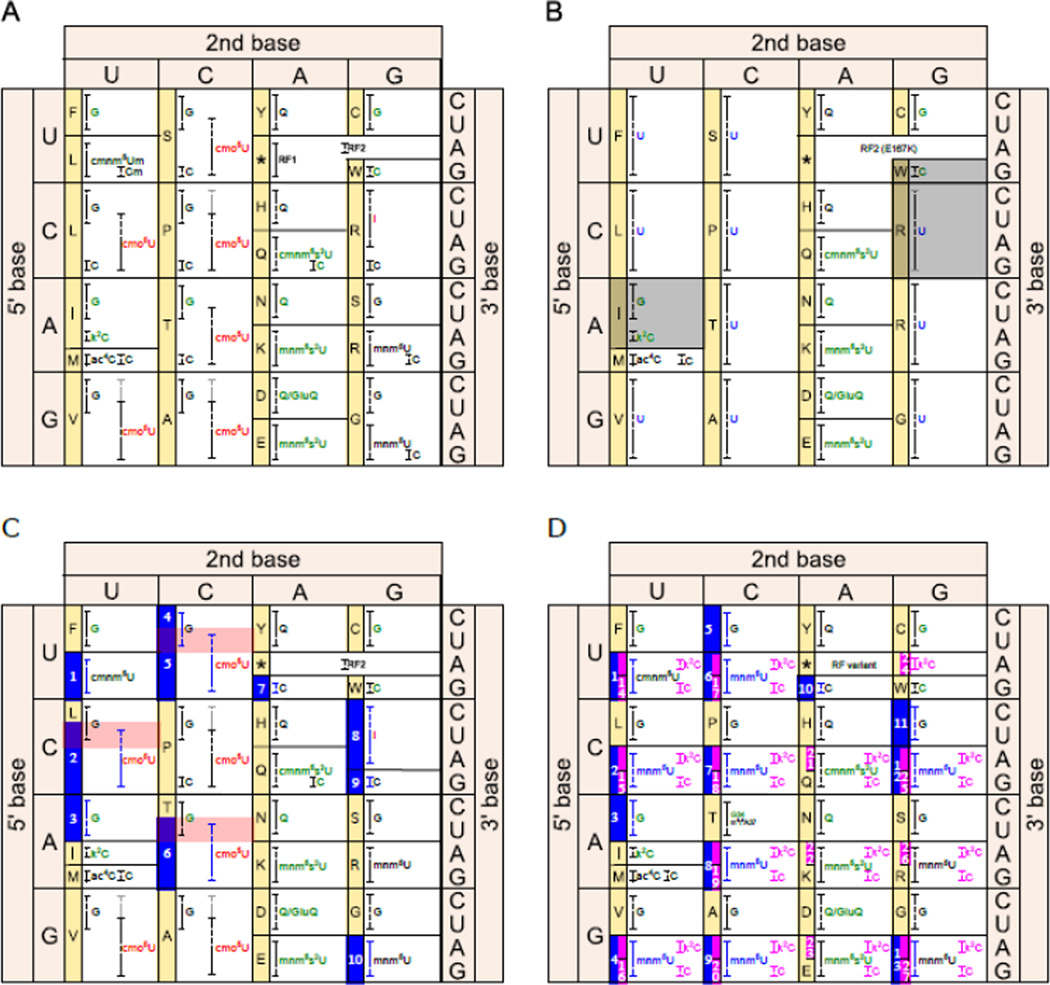
Figure 4. Minimal and maximal genetic codes using triplet codons composed of four nucleotide types (U, C, A, G).
The proposed genetic codes are one possible permutation representing several possible ways to reassign redundant codons. Dashed brackets represent codon recognition ranges for a given anticodon: black is codon recognition agreeing with wobble rules [192,193]; gray is empirical data that supersedes wobble rules [94]; blue and magenta are proposed new tRNAs that can be assigned to nsAAs. Labels correspond to the wobble nucleotide at tRNA position 34 (cmo5U = uridine 5-oxyacetic acid, mnm5U = 5-methylaminomethyluridine, cmnm5U = 5-carboxymethylaminomethyluridine, cmnm5Um = 5-carboxymethylamino-methyl-2′-O-methyluridine, mnm5s2U = 5-methylaminomethyl-2-thiouridine, cmnm5s2U = 5-carboxymethylamino-methyl-2-thiouridine, I = inosine, k2C = lysidine, Q = queuosine, GluQ = glutamylqueuosine) [173]. Green letters indicate natural tRNA identity determinants that may be difficult to change without disrupting aminoacylation. Red letters indicate natural anticodon modifications that increase anticodon promiscuity. Blue and magenta letters represent proposed changes in the tRNA wobble position that would alter codon recognition. Amino acid assignments are indicated in the yellow sidebars. M refers to both Met and fMet (translation initiation). Codons available for new amino acids are indicated by blue and magenta boxes with white numbers. Selenocysteine is not shown. (A) The E. coli genetic code is presented based on Björk et al. [94] and tRNA identity determinants are from Giegé et al. [110]. All 64 codons are used to encode 20 amino acids. (B) A minimal genetic code utilizing all 64 codons would require initiation at AUG, one release factor (RF2 E167K mutants can terminate all 3 stop codons [98]), and one tRNA for each of the 20 amino acids. Unmodified uracils in the wobble positions would allow tRNAs to recognize all codons in a family group, allowing redundant tRNAs to be deleted. Gray shaded boxes represent additional anticodons that could be potentially deleted (tRNAArgUCG would encode the same amino acid as tRNAArgUCU and it may be possible to remove Ile [103] and Trp [104] from the genetic code). Conveniently, the wobble nucleotide is rarely a tRNA identity determinant [110]. The two relevant exceptions, tRNAPheGAA (G34) and tRNAGluUUG (cmnm5s2U34), are weak identity determinants [110], so the proposed changes may be tolerated by their respective aaRSs. (C) The genetic code can be expanded to provide 7 unambiguous and 3 ambiguous anticodons by simply deleting tRNAs and introducing orthogonal aaRS/tRNA pairs encoding new amino acids. This analysis assumes that the original aaRS/tRNA identity determinants/antideterminants can be overcome by a metagenomic search for an orthogonal aaRS/tRNA pair and subsequent directed evolution to optimize their orthogonality. Red shaded boxes represent the three codons that would gain ambiguous translation function upon introduction of an orthogonal aaRS/tRNA pair. GUN, GCN, and CCN were not included, since the cmo5U-containing anticodon has been empirically shown to recognize all four codons in their respective families [94]. The UAG codon has already been reassigned [13]. (D) Replacing the cmo5U and inosine wobble nucleotides with mnm5U nucleotides could liberate 13 unambiguous anticodons for reassignment (33 total amino acids; changes indicated in blue). Doing so would require the inactivation cmoB [105] and the engineering of mnmE and mnmG to recognize additional tRNAs [106,107]. Taken a step further, the maximal genetic code would have unique amino acid assignments for all NNA and NNG codons (NNA: engineer tilS [108] to lysidinylate additional tRNAs so that they only base pair with A; NNG: change the tRNA wobble bases to cytosine so that they only base pair with G). Anticodon modifications capable of splitting NNY codons into unambiguous NNU and NNC codons have not been reported, but such modifications have not been ruled out. Additionally, it may be possible to engineer a release factor to terminate translation only at UAA codons, thereby liberating both UAG and UGA codons. The proposed changes would liberate 27 unambiguous anticodons (47 total amino acids; changes indicated in magenta). This strategy may require directed evolution to overcome the tRNA identity determinants for Glu, Gln, and Lys [110]. Another potential complicating factor is that G + C anticodon content may affect cognate and near-cognate decoding efficiencies, just as the G + C rich anticodons for Val, Ala, and Pro break the wobble rules [94].