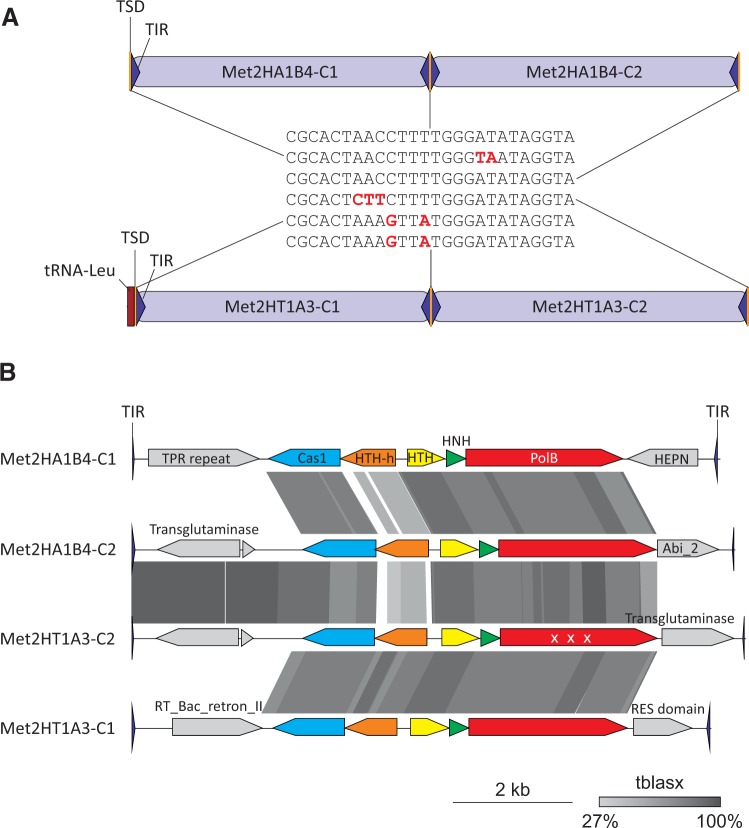
Fig. 2.—
Comparison of tandemly integrated casposons in different Methanosarcina species. (A) Schematic representation of the genomic loci within Methanosarcina sp. 2.H.A.1B.4 (top) and Methanosarcina sp. 2.H.T.1A.3 (bottom) containing casposons integrated into the intergenic region and tRNA-Leu gene, respectively. TIR, TSD, and tRNA gene are depicted with blue triangles, yellow bars, and a red rectangle, respectively. The sequences of the corresponding TSDs are shown in the middle and substitutions with respect to the top sequence are highlighted in red. (B) Comparison of the genome maps of casposon depicted in panel A. Pairwise tBLASTx hits between casposons are indicated by different shades of gray (the identity scale is included). “X X X” indicates that the gene is fragmented in Met2HT1A3-C2. TPR, tetratricopeptide; HTH-h, helix-turn-helix protein with a C-terminal HEAT repeat domain; RT_Bac_retron_II, retrotransposon/retron-like reverse transcriptase; PolB, family B DNA polymerase; HNH, HNH endonuclease.