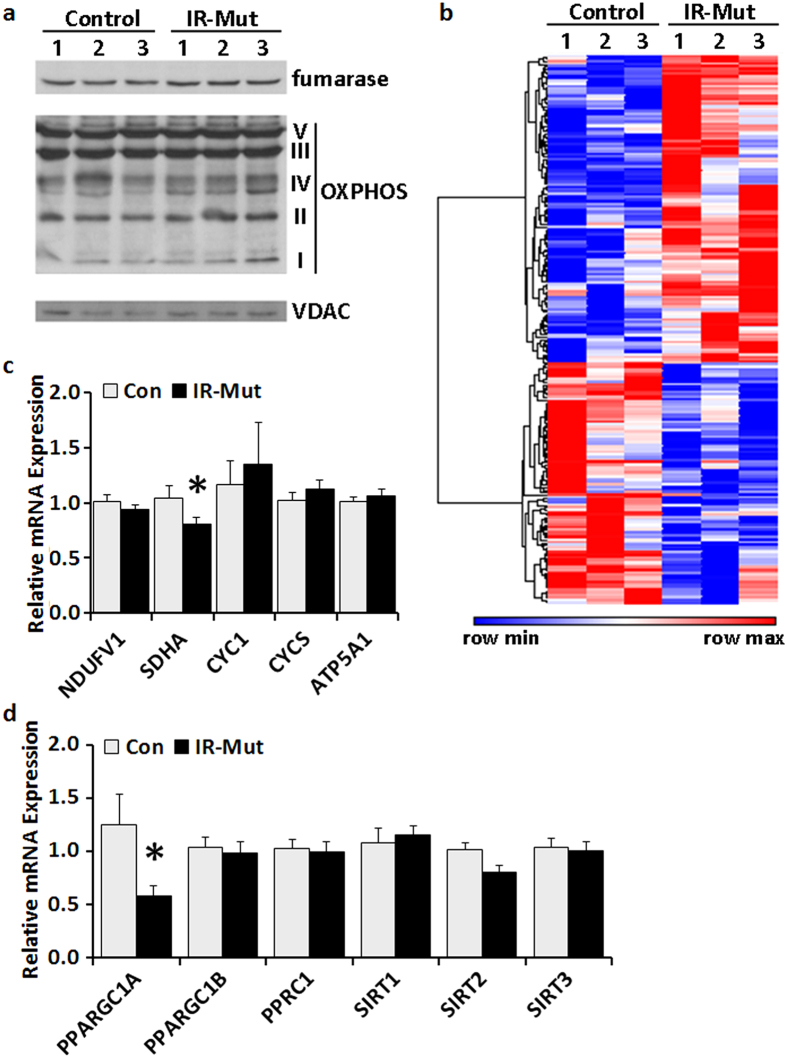
Figure 3. IR-Mut iPSC have decreased PGC1α expression.
(a) Representative western blots showing protein expression of fumarase and key enzymes in OXPHOS. VDAC was used as a loading control (n = 3). (b) Heat map of differentially expressed genes (p < 0.05) related to mitochondria using ontology terms “mitoc*” as determined by microarray analysis, with red indicating increased expression and blue indicating decreased expression. (c) Relative mRNA expression of selected nuclear-encoded mitochondrial genes, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control (n = 3). (d) Relative mRNA expression of selected genes regulating mitochondrial biogenesis and metabolism, as assessed by qRT-PCR. Data are normalised to 36B4 and expressed relative to control (n = 3). *indicates p < 0.05 for IR-Mut vs. control.