Figure 3. Identification of Sirt5 substrates.

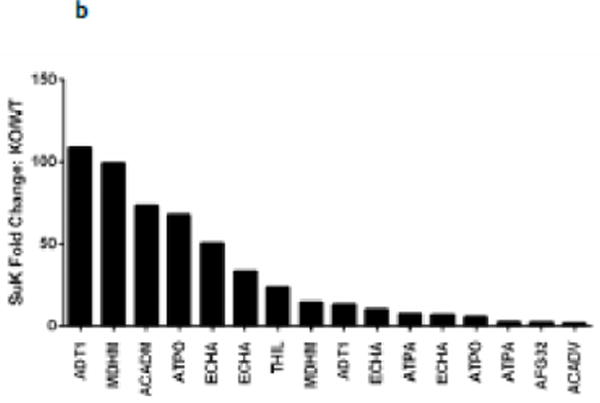
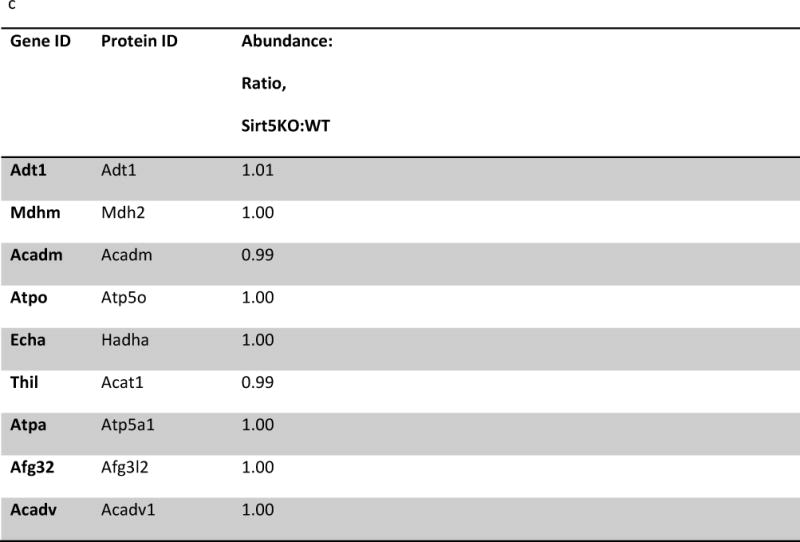
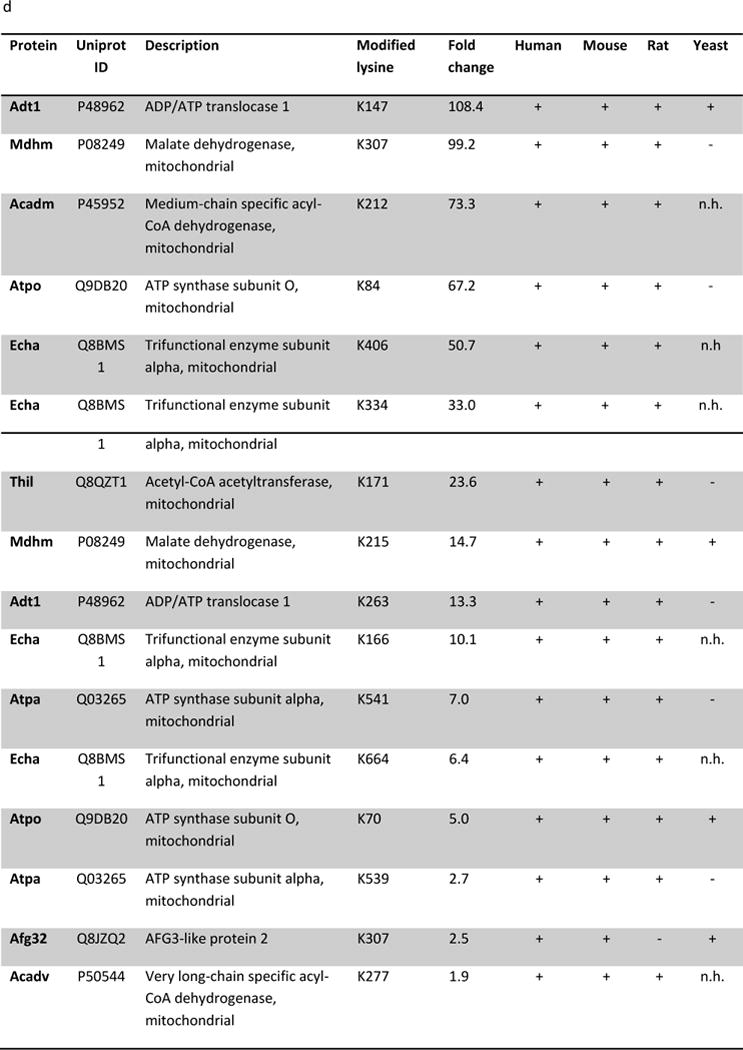
(a) IPA core analysis was used to functionally classify proteins mapped from peptides that were detected only Sirt5−/− samples. (b) A label free analysis program (QUOIL) was used to determine the relative abundance of succinylated peptides detected in both WT and Sirt5−/− samples. Represented here are the 16 peptides from 9 proteins increased by > 30% with p < 0.05 in Sirt5−/− hearts. (c) Relative protein quantitation was calculated based on the intensities of TMT report ions using Scaffold 4.0 (Proteome Software, Portland, OR). The average ratio of protein expression (Sirt5−/−: WT) is expressed numerically. The data demonstrate no significant differences in total protein abundance between Sirt5−/− and WT mice. (d) Conservation of the Sirt5 substrate lysines across human, mouse, rat, and S. Cerevisiae (yeast). Protein alignments were performed with the “Align” tool at www.uniprot.org; “+” indicates a conserved lysine, “−“ indicates that the lysine is not conserved, “n.h.” indicates that a homologous protein is not expressed by the organism.
