Abstract
We identified a homologue of the molluscan acetylcholine-binding protein (AChBP) in the marine polychaete Capitella teleta, from the annelid phylum. The amino acid sequence of C. teleta AChBP (ct-AChBP) is 21–30% identical with those of known molluscan AChBPs. Sequence alignments indicate that ct-AChBP has a shortened Cys loop compared to other Cys loop receptors, and a variation on a conserved Cys loop triad, which is associated with ligand binding in other AChBPs and nicotinic ACh receptor (nAChR) α subunits. Within the D loop of ct-AChBP, a conserved aromatic residue (Tyr or Trp) in nAChRs and molluscan AChBPs, which has been implicated directly in ligand binding, is substituted with an isoleucine. Mass spectrometry results indicate that Asn122 and Asn216 of ct-AChBP are glycosylated when expressed using HEK293 cells. Small-angle X-ray scattering data suggest that the overall shape of ct-AChBP in the apo or unliganded state is similar to that of homologues with known pentameric crystal structures. NMR experiments show that acetylcholine, nicotine, and α-bungarotoxin bind to ct-AChBP with high affinity, with KD values of 28.7 μM, 209 nM, and 110 nM, respectively. Choline bound with a lower affinity (KD = 163 μM). Our finding of a functional AChBP in a marine annelid demonstrates that AChBPs may exhibit variations in hallmark motifs such as ligand-binding residues and Cys loop length and shows conclusively that this neurotransmitter binding protein is not limited to the phylum Mollusca.
Nicotinic acetylcholine receptors (nAChRs)1 and the structurally related GABAA and GABAC, serotonin 5-HT3, and glycine receptors are well-studied members of the Cys loop ligand-gated ion channel superfamily in the central and peripheral nervous system (1, 2). These receptors are comprised of homo- or heteropentamers of homologous subunits, with an extracellular N-terminal ligand binding domain. In the presence of an agonist, these receptors undergo a conformational change in the extracellular domain that leads to channel opening in the pore domain (3). Each subunit can be subdivided into two principal domains: extracellular and transmembrane. The extracellular domain (ECD) carries the acetylcholine (ACh) binding site at the subunit interface, and the transmembrane ion pore domain (IPD) delineates an axial cation-specific channel (4, 5). These topologically distinct domains are allosterically coupled to each other. Consequently, nAChRs possess the structural elements necessary to convert a chemical signal, typically a local increase in extracellular ACh concentration, into an electrical signal mediated by the opening of the ion channel.
The three-dimensional structure of the Cys loop receptor ECD was unknown until the identification and structure determination of the acetylcholine-binding protein (AChBP) from the snail Lymnaea stagnalis [ls-AChBP (6)]. AChBP is a pentameric protein that is secreted from glia cells in the central nervous system of this freshwater snail, where it has been proposed to modulate synaptic transmission, although a recent study raises doubts about the synaptic localization of AChBP in vivo (7). AChBP was then subsequently identified in the molluscs Aplysia californica [ac-AChBP (8)] and Bulinus truncatus [bt-AChBP (9)]. All three of these proteins come from organisms within the same class of mollusc, namely Gastropoda. Recently, another mollusc, the abalone (Haliotis discus hannai), was shown to have a functional homologue of AChBP (10). Notably a homologue of AChBP from the pearl oyster Pinctada fucata, involved in the control of the morphology of nacre lamellae, appears to bind calcium carbonate (11). To date, the only isolated and functionally characterized AChBPs have come from molluscs. This trend has raised questions about the existence of nonchannel homologues of AChBP throughout the other phyla of metazoans, such as the protostome phyla Arthropoda and Annelida. The presence of an AChBP in specific phyla (but not others) may shed light on phylum-specific mechanisms of regulating neurotransmitter levels, or the evolutionary history of the protein. Furthermore, the identification of AChBPs in other phyla besides mollusk and analysis of their sequence and structure will provide information about critical residues for agonist binding of ACh or other molecules of therapeutic medical interest, such as nicotine. The finding of a functional AChBP in the annelid phylum, such as the one we have reported here, adds a new chapter to the study of AChBPs and broadens what is known about the distribution of AChBPs in the animal kingdom.
EXPERIMENTAL PROCEDURES
Identification of ct-AChBP and Homology Modeling
The cDNA encoding the Capitella teleta homologue of AChBP (GenBank accession number EY637248) was a kind gift from R. Savage (Williams College, Williamstown, MA). A His tag and V5 epitope were appended to the C-terminus of the protein. CHO cells were transfected and harvested using TALON cobalt beads. A homology model for the C. teleta AChBP (ct-AChBP) was generated using the SwissModel server using Protein Data Bank (PDB) entry 1I9B for the L. stagnalis AChBP as the query. A dimer was created using the SwissModel server, and the ligand ACh was inserted on the basis of its predicted location in the ligand binding pocket in our α7 nAChR model (PDB entry 1OL4) after structural alignment with our ct-AChBP model.
Protein Purification
The gene for AChBP from C. teleta was cloned into vector pBUDCE4.1 (Invitrogen); the gene was inserted using the KpnI and XhoI sites, which were introduced by PCR. The stop codon was removed during the polymerase chain reaction (PCR) so that the C-terminus would acquire the V5 and His tags from the vector for Western identification and purification, respectively. The pBUDCE4.1-AChBP vector was transfected into HEK293-f cells grown in DMEM (GIBCO) containing 10% fetal bovine serum (Hyclone). A stable cell line was selected using 400 μg/mL Zeocin (Invitrogen). Once established, the HEK293-f-AChBP cells were adapted for growth in Freestyle 293 medium as a suspension culture in the presence of Zeocin. Cells were removed from the conditioned medium by centrifugation at 342g for 5 min. The medim was then clarified by centrifugation at 34000g for 10 min. The His-tagged AChBP was batch loaded overnight onto 2 mL of cobalt Talon matrix (Clontech) per liter of medium in the presence of 10 mM imidazole (Sigma) at 10 °C. The Talon matrix was collected and packed into a 10 mm × 20 cm Bio-Rad econo-column. The Talon column was washed with 3 column volumes of wash buffer [50 mM Hepes (pH 7.5), 300 mM NaCl, and 10 mM imidazole], and the protein was eluted with wash buffer containing 250 mM imidazole. Fractions (1 mL) were collected during the elution, and all fractions containing protein as determined by the Pierce Coomassie protein assay were pooled and concentrated for injection onto a Suderdex 200 column (GE Healthcare). The Superdex 200 column was developed using PBS buffer [25 mM potassium phosphate (pH 7.5) and 150 mM NaCl]. Approximately 1 mg of pure AChBP was produced per liter of cultured medium used.
Mass Spectrometry
Gel bands were excised manually and digested with trypsin (Promega), chymotrypsin, or GluC (both from Roche) for 8 h in an automated fashion with a Progest robotic digester (Genomic Solutions). LC–ESI-MS was performed using a nanoAcquity UPLC system in-line with a Waters-Micromass Q-Tof Premier mass spectrometer (Waters). Chromatographic analyses were performed using a 3 μm, 100 μm × 100 mm, Atlantis dC18 column (Waters) and a flow rate of 300 nL/min. A C18 trapping column (180 μm × 20 mm, 3 μm particle size) was positioned in-line of the analytical column and upstream of a microtee union used as a junction and a vent for trapping. Trapping was performed for 3 min at a flow rate of 5 μL/min, using the initial solvent composition. Briefly, a 2 μL aliquot of the AChBP in-gel digest was loaded onto the column. Peptides were eluted using a linear gradient from 98% solvent A [0.1% formic acid in water (v/v)] and 2% solvent B [0.1% formic acid in acetonitrile (v/v)] to 40% solvent B over 90 min. Mass spectrometer settings for the MS analysis were as follows: capillary voltage of 2.8 kV, cone voltage of 20 V, collision energy of 5.0 V, and source temperature of 80 °C. The mass spectra were recorded over the mass range of 200–2000 Da. MS/MS data were acquired in a data-dependent mode, using collision energies based on the mass and charge state of the candidate ions or a collision energy ramp from 30 to 40 V. The mass spectrometer was externally calibrated using the fragment ions of Glu-fibrinogen peptide 23.
Nuclear Magnetic Resonance
The ligand binding characteristics of the putative AChBP were surveyed using NMR saturation transfer difference (STD) experiments in which various neurotransmitters (100 μM) were evaluated (12, 13). Samples contained 10 μM AChBP in 100% D2O and 25 mM phosphate (pH 7.0). All NMR samples used either the reference 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) (Cambridge Isotope Laboratories, Andover, MA) or d6-trimethylsilylpropionate (TMSP) (Isotec, Miamisburg, OH) for chemical shift and concentration. All experiments were performed on a 500 MHz Varian INOVA NMR spectrometer with a cold probe. Each line broadening experiment was conducted using Varian’s presat experiment with a spectral width of 14.0 ppm, 14K points, and 64 scans. The STD experiments were conducted using Varian’s satxfer1D experiment, with 0.88 ppm as the on frequency, 30.14 ppm as the off frequency, and an acquisition time of 0.5 s. The survey STD experiments for determining binding qualitatively used 1K scans. The ligand competition experiments used 1K scans for nicotine, 4K scans for α-bungarotoxin, and 16K scans for acetylcholine.
The choline dissociation constant was calculated on the basis of measurements of shift or line width as a function of the choline concentration, which was assumed to vary proportionally with the fraction of bound choline according to
| (1) |
where PT is the total protein monomer concentration, LT is the choline concentration, and δ0 and δB are the uncomplexed and protein-complexed values, respectively, for the shift or line width.
The binding affinities of nicotine, acetylcholine, and α-bungarotoxin were determined using STD displacement (12, 13). The ligand competition experiment extends the range of KD values accessible to the STD experiment (14). Because of the overlap of the N(CH3)3 peaks of choline and acetylcholine, the choline CH2 resonances were used as the displacement probes upon addition of acetylcholine, while the N(CH3)3 STD peak intensity of acetylcholine was used in the nicotine titration studies. The NCH3 resonance of nicotine was used in the α-bungarotoxin ligand competition study. The data were analyzed utilizing the nonlinear least-squares fitting subroutines of Mathematica (Wolfram Research, Champaign, IL) based on the expression given by Wang (15) for the fraction of reporter ligand bound in the presence of a second, competing ligand, L2:
| (2) |
with pB(L1), as defined in eq 1, given by
and
| (3) |
where
| (4) |
where L1T and L2T are the total concentrations of the reporter and competing ligand, respectively, KD1 and KD2 are the corresponding dissociation constants, and PT is the total protein concentration (15). To determine the binding constant of acetylcholine, a sample containing 100 μM choline and 5 μM AChBP was titrated with acetylcholine in the appropriate concentration range. Since choline was determined to be stoichiometrically displaced by nicotine as a result of the large difference in the dissociation constants, it was found to be unsuitable for the determination of the nicotine KD value; to obtain sufficient nonlinearity, acetylcholine (100 μM) was used as the reporter ligand for determination of the nicotine KD value. The α-bungarotoxin binding constant was determined by the displacement of 50 μM nicotine using 5 μM AChBP. The standard errors of the estimated KD values of the parameters of the model were derived using a standard first-order asymptotic covariance matrix (16).
SAXS Data Acquisition and Processing
SAXS (small-angle X-ray scattering) data were collected at beamline X9 at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY). The wavelength of the beam was 0.85 Å. Protein was dialyzed into a 10 mM phosphate/300 mM NaCl buffer (pH 7.0) for SAXS analysis and was treated with PNGase F to remove any N-linked glycans. Protein and buffer samples were exposed for 90 s at 25 °C in a quartz flow cell at a flow rate of 28 μL/min. The recorded intensities of the sample were circularly averaged and scaled to produce a relative scattering intensity (I) as a function of momentum transfer vector, q [q = (4π sin θ)/λ], after subtraction of buffer scattering contributions. SAXS data were collected for three protein concentrations [3.8, 2.85, and 1.9 mg/mL for the apoprotein (Figure 1 and Figure S1 of the Supporting Information)]. Background subtraction, averaging, and scaling were conducted using the Primus software package (17). The three-dimensional shape of ct-AChBP was constructed from the SAXS data using GASBOR22IQW (18), by calculating the distribution of linearly connected 1.9 Å spheres that best fit the scattering data. Each calculation was repeated at least five times with different random starting points for the Monte Carlo optimization algorithm. A P5 symmetry constraint was used; in the absence of this restraint, models did not consistently converge to similar structures. From these runs, the predicted structure with the lowest deviation of the calculated scattering profile from experimental data was used for interpretation. CRYSOL (19) was used to compare the SAXS-based models with the atomic structure obtained from X-ray crystallographic data, and the structures were superimposed using SUPCOMB13 (20).
FIGURE 1.
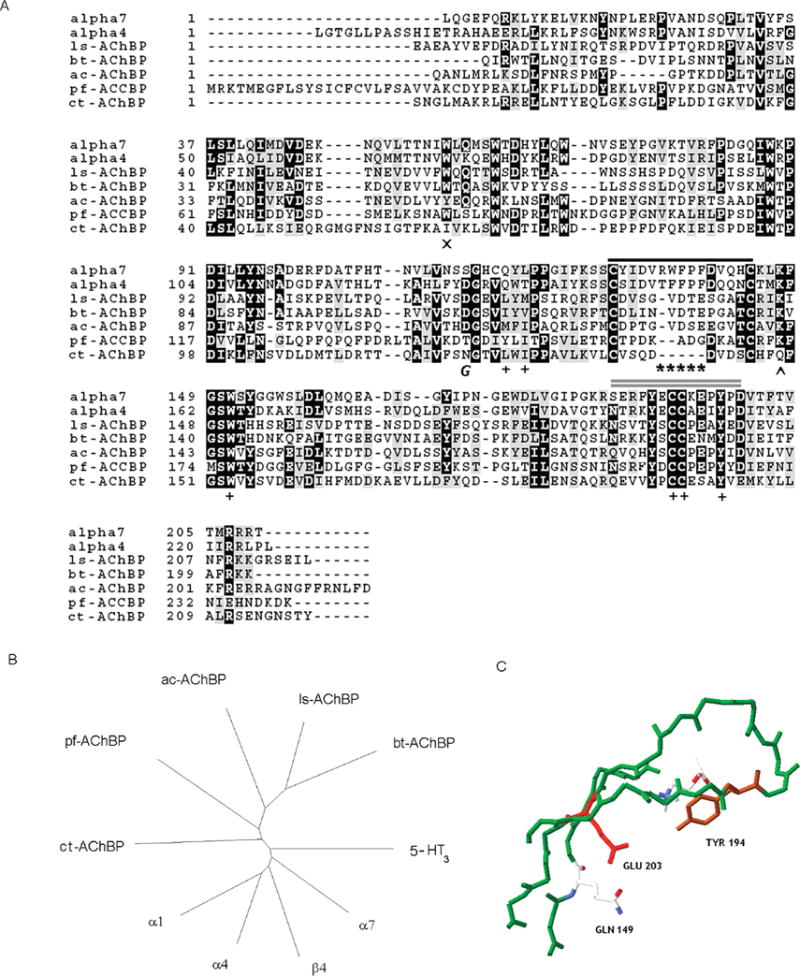
(A) Amino acid alignment of ct-AChBP with other AChBPs and the extracellular domain of nAChR subunits (human α4 and α7). The shortened Cys loop of AChBP is highlighted by asterisks. Residues deemed important for agonist binding are designated with a plus sign; a single black horizontal line lies atop the Cys loop, and a double gray horizontal line lies atop the C loop. The glycosylated Asn122 is designated by an italic letter G, and the caret designates the position of Gln149. The AChBPs listed are ls-AChBP from L. stagnalis, bt-AChBP from B. truncatus, ac-AChBP from A. californica, pf-AChBP from P. fucata, and ct-AChBP from C. teleta. (B) Radial (neighbor-joining) tree based on a Clustal alignment of amino acid sequences of indicated Cys loop receptors. (C) Variation of a conserved amino acid triad found in ct-AChBP. The C loop region from a homology model of ct-AChBP is colored green, Tyr194 brown, Glu203 red, and Gln at position 149 in CPK format.
RESULTS
Identification of the C. teleta AChBP
Novel AChBPs have been identified using an in silico homology-based screening approach. We took advantage of the fact that the known molluscan AChBPs have a much more hydrophilic Cys loop than nicotinic ACh receptors (nAChRs), an expected property given that the Cys loop of AChBP does not interact with the hydrophobic transmembrane regions that are not present in AChBP. We identified a novel AChBP candidate from C. teleta, a marine polychaete known to be tolerant of stressful conditions, and often found in polluted waters. The C. teleta AChBP homologue (ct-AChBP) was identified by performing a BLAST search (tBLASTN) against the non-rat, non-human EST (Expressed Sequence tag) nucleotide database at NCBI using the L. stagnalis AChBP as the query sequence.
A hydrophobic signal sequence on the N-terminus of the ct-AChBP clone (GenBank accession number EY637248) was observed: the Signal Sequence prediction server SignalP 3.0 predicts that a 217-amino acid protein is secreted after cleavage. This sequence is shown in the alignment in Figure 1A. The ct-AChBP sequence was 28–30% identical to those of the molluscan AChBPs from Lymnaea and Aplysia but only 21% identical to that of the Bulinus AChBP and 25% identical to that of the rat α7 nAChR. The radial tree showing the relationships among the various known AChBPs and nAChRs is shown in Figure 1B. Of particular note is the fact that the ct-AChBP sequence has only eight amino acids between the cysteines of the Cys loop, the shortest of any AChBP or Cys loop receptors identified to date. In addition, ct-AChBP has a variation of a conserved Lys-Tyr-Asp triad (in the β7, β9, and β10 strands, respectively) near the C loop which is associated with structural changes that accompany ligand binding (21). Although the Tyr residue at position 194 is conserved in ct-AChBP and a negatively charged residue (glutamic acid rather than aspartic acid) is present at position 203, position 149 in ct-AChBP has a Gln residue rather than a charged Lys residue (Figure 1C). This Gln residue would not be expected to form the salt bridge with the nearby acidic residue associated with the closed state (3).
Protein Purification
Efficient secretion of the processed protein expressed in HEK293 cells was demonstrated via Western blotting using an antibody against the V5 epitope (Figure 2B) and confirmed by mass spectral analysis. The ct-AChBP band, which runs at 35–37 kDa on the SDS gel, is much larger than the expected 28.9 kDa band from the cleaved protein, including the C-terminal tags from the vector. This suggests that during processing of the correctly folded protein, it is post-translationally modified. The secreted protein was captured by the cobalt Talon matrix and pulse eluted using 250 mM imidazole (Figure 2A). We then subjected the virtually pure ct-AChBP to size exclusion chromatography to remove minor contaminants and to exchange it into a suitable buffer for further study (Figure 2A). A homology model of ct-AChBP structurally aligned with the α7 nAChR model was created using the SwissModel server (Figure 2C).
FIGURE 2.
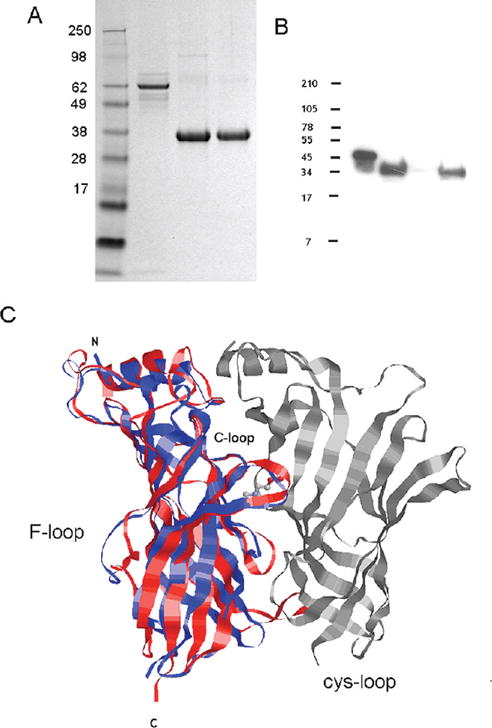
(A) Coomassie-stained SDS–PAGE gel of ct-AChBP: lane 1, SeeBlue Plus 2 molecular mass marker; lane 2, medium from ct-AChBP-transfected HEK293 cells; lane 3, pooled fractions of ct-AChBP after elution from the cobalt Talon matrix; and lane 4, pooled fractions of ct-AChBP after the Superdex 200 column. A precast NuPAGE 4–12% Bis-Tris precast gel was run using NuPAGE MES SDS running buffer. (B) Western blot analysis of ct-AChBP protein using anti-V5 antibody: lane 1, positive control, BSA; lane 2, medium from COS cells; lane 3, medium from CHO cells; and lane 4, medium from HEK293 cells. A volume of 20 μL was added to each lane. (C) Homology model of ct-AChBP dimer (red) superimposed on the dimer of a rat α7 nAChR model (blue; PDB entry 1OL4). Proteins are shown in ribbon form; both are colored gray on the negative face. ACh (stick form) is shown in its predicted position near the C loop. The C loop, F loop, Cys loop, and N- and C-termini are labeled accordingly.
Mass Spectrometry
To characterize the mammalian expressed AChBP, MALDI-ToF and nanoLC-ESI-QToF mass spectrometry were employed. Analysis of intact, purified AChBP by MALDI-ToF mass spectrometry yielded a broad, ill-defined peak (Figure S2A of the Supporting Information), suggesting the possibility of a heavily adducted or heterogeneous sample. The centroid mass of the AChBP ion was approximately 33.5 kDa, but the predicted mass of the expressed AChBP (with the C-terminal vector sequence, including V5 and His tags) is only 28.9 kDa. This disparity between the theoretical and observed masses and the poorly defined nature of the peak suggested that the sample could be glycosylated. To test this, the sample was treated with PNGase F to remove any N-linked glycans. Upon PNGase treatment, three distinct bands, suggesting two sites of glycosylation, could be visualized by SDS–PAGE, whereas a single band was observed without PNGase treatment (Figure S2B of the Supporting Information). In efforts to identify the types and locations of the glycans or other posttranslational modifications, AChBP was digested from the gel with a variety of enzymes. The proteolytic digests were analyzed by nanoLC-ESI-MS and MS/MS and yielded 83% sequence coverage. Interestingly, MS/MS data were obtained on a doubly charged ion at m/z 360.69, and an extensive y-ion series identified the peptide as the SNGLMAK sequence (Figure S2C of the Supporting Information). While this peptide is not a strictly predicted tryptic peptide of AChBP as expressed, it does match a tryptic peptide of the expected mature form of AChBP as the 20 N-terminal residues are predicted with a 98% probability by Signal P software to be proteolytically removed, leaving Ser21 as the new N-terminus, when the protein is secreted. Additionally, MS/MS data of a doubly charged ion at m/z 938.55 indicated a disulfide bond between cysteines 196 and 197 (Figure S2D of the Supporting Information). Finally, glycosylation at Asn122 was also observed. Asn122 is a consensus site for glycosylation following the NX(S/T) motif. The glycan at this site is heterogeneous with the most abundant species being the A1, A1F, A2, and A2F types of complex glycans (Figure 3A). The site and nature of the glycan were confirmed by MS/MS. As an example of these data, the MS/MS spectrum of a triply charged ion at m/z 1536.07 is shown in Figure 3B. This ion corresponds to residues 112–132 with an A2F type glycan occupying Asn122. AChBP contains a second consensus site for glycosylation at Asn216. An ion at m/z 558.2 with an elution time of approximately 30 min could be readily observed in the PNGase F-treated sample, but not in the untreated samples (Figure 3C). As determined by MS/MS, this ion was found to correspond to SENGDSTYSR, where Asn216 is replaced with aspartic acid (in italics) upon glycan cleavage by PNGase F (Figure 3D). Even though the nature of the glycan at this site could not be determined, the substitution of the asparagine with aspartic acid upon PNGase F treatment confirms Asn216 as a second site of glycosylation. The location of Asn122 in the E-loop is shown in the ct-AChBP dimer model in Figure 3E.
FIGURE 3.
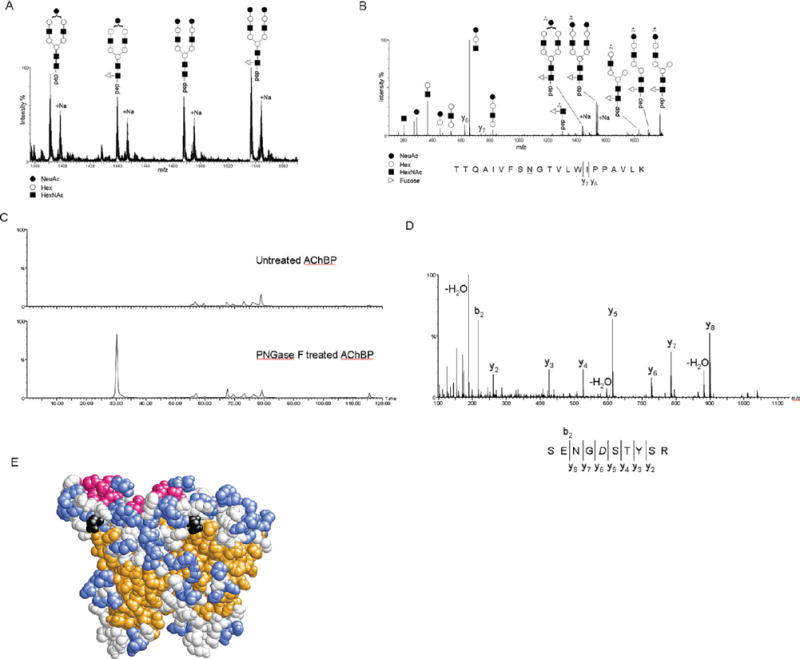
(A) Representation of the m/z range from 1380 to 1560 of an MS spectrum from a nanoLC-ESI-MS experiment and MS/MS analysis of the tryptic digest of ct-AChBP. Triply charged ions of the T17 peptide with Asn122 modified with the A1 (m/z 1390.34), A1F (m/z 1439.02), A2 (m/z 1487.34), and A2F (m/z 1536.07) types of complex glycans and the respective sodiated adducts. (B) Representation of the MS/MS spectrum of the A2F-modified T17 peptide from ct-AChBP. Generally, the CID-based fragmentation mechanism provides mostly information about the glycan structure. This can be seen by the extensive fragmentation of the glycan into its constituent mono-, di-, and trisaccharides, as well as pieces of the complex glycan remaining on the peptide. In this instance, MS/MS also provided sparse information about the peptide itself by the presence of the y6 and y7 ions. (C) Extracted ion chromatogram for the ion at m/z 558.2 for the untreated (top) and PNGase F-treated (bottom) ct-AChBP. The presence of this ion in only the PNGase F-treated ct-AChBP sample confirms Asn216 as a site of N-linked glycosylation. (D) MS/MS spectrum of the ion at m/z 558.2 that, via an extensive y-ion series, confirms the sequence of the peptide as SENGDSTYSR, where the italic D denotes the former site of glycosylation. (E) Space-filling model of the ct-AChBP homology model dimer. The predicted β-strands are colored orange, α-helices red, and loops and coils colored blue and white, respectively. The asparagine residue at position 122 is colored black.
Nuclear Magnetic Resonance
Ligand binding preferences of the putative ct-AChBP were initially characterized by saturation transfer difference (STD) experiments using a mixture of neurotransmitters, each present at 100 μM (Figure 4). From these data, it was clear that the ct-AChBP protein exhibits the binding characteristics expected for an AChBP, with affinity for choline, acetylcholine, and nicotine, but not for dopamine, arginine, or glutamate. A comparison of the resonance intensities (Figure 4B,D) supported the qualitative relations: KD(choline) > KD(acetylcholine) > KD(nicotine). The relatively low affinity for choline facilitated the direct determination of its KD value by direct titration, in which either the shift or the line width of the N(CH3)3 resonance was monitored. The KD determined using this approach is dependent on the number of choline binding sites of the AChBP, and it was initially assumed that there is one binding site per monomer, or five equivalent binding sites in the pentamer. A fit of the chemical shift titration data (Figure 4E) yielded a KD of 153 ± 49 μM and a shift change for the choline N-methyl protons in the bound state of −0.46 ppm. Since in the reported crystal structure of the AChBP–carbamylcholine complex (PDB entry 1UV6) not all of the binding sites are occupied, the calculation was repeated assuming only a single binding site per pentamer. This assumption resulted in a moderate increase in the calculated KD, and a predicted shift for the bound N-methyl protons of −2.65 ppm. This extremely large upfield shift would be unusual, particularly since it represents an average for three methyl groups. Hence, the shift data are more consistent with the single binding site per monomer model. Analysis of the choline N-methyl line width (Figure 4F) as a function of choline concentration yielded a similar KD value of 173 ± 56 μM, for a mean KD for choline of 163 ± 37 μM. The dependence of the shift on the protein concentration shown in Figure 4 is consistent with a ligand in fast exchange. This conclusion is also supported by the relatively high dissociation constant. For a typical diffusion-controlled binding interaction of a small protein ligand with a ka of ~108 M−1 s−1 (22–24), the value given above corresponds to a dissociation rate constant k−1 of 1.6 × 104 s−1, consistent with the fast exchange limit.
FIGURE 4.
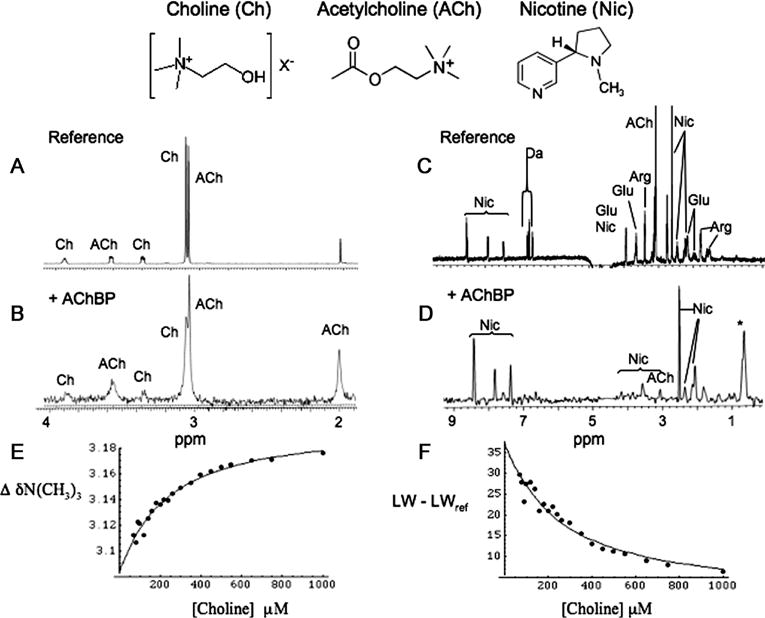
NMR characterization of ct-AChBP ligand binding. (A) Expanded region of a reference 1H spectrum of 100 μM acetylcholine (ACh) and choline (Ch). (B) STD spectrum after the addition of 10 μM ct-AChBP. As is apparent from the peak intensity differences, the STD intensities for acetylcholine are greater than those of choline. (C) Reference 1H spectrum of acetylcholine (ACh), glutamic acid (Glu), dopamine (DA), arginine (Arg), and nicotine (Nic). (D) STD spectrum after addition of 10 μM ct-AChBP, revealing the dominant nicotine resonances, and smaller acetylcholine resonances. Resonance assignments for the aromatic nicotine peaks and the acetylcholine methyl groups are indicated. The asterisk denotes the on-resonance saturation frequency. 1H shift (E) and line width (F) of the choline N(CH3)3 resonance as a function of choline concentration in the presence of 50 μM ct-AChBP. The buffer used in the NMR studies was 25 mM phosphate (pH 7.0) in 100% D2O.
Dissociation constants for acetylcholine (Figure 5A), nicotine (Figure 5B), and α-bungarotoxin (Figure 5C) were then determined by ligand competition studies, and the data were analyzed as described in Experimental Procedures. For the acetylcholine KD determination, choline was used as the reporter ligand; for the nicotine KD, acetylcholine served as the reporter ligand, and for the α-bungarotoxin study, the nicotine resonance was observed. Fitting the data as described in Experimental Procedures yielded KD values of 28.7 ± 1.7 μM, 209 ± 51 nM, and 110 ± 13 nM, respectively.
FIGURE 5.
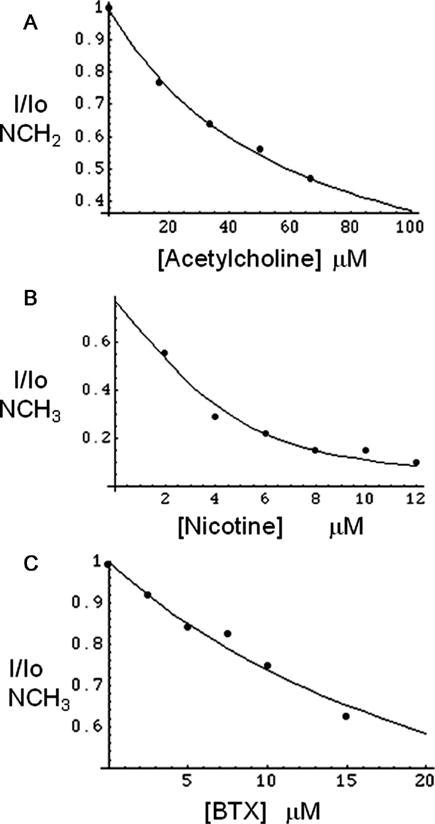
STD displacement titrations of (A) acetylcholine using the choline N(CH2) resonance (choline concentration of 100 μM and ct-AChBP concentration of 5 μM), (B) nicotine using 100 μM acetylcholine as the NMR reporter ligand, and (C) α-bungarotoxin (BTX) using 50 μM nicotine as the NMR reporter ligand. NMR studies were performed at 25 °C.
Small-Angle X-ray Scattering (SAXS)
AChBP (treated with PNGase F) was analyzed with small-angle X-ray scattering (Figure 6) in the absence of ligand (i.e., apo form). Guinier plots for the apoprotein are linear over a q range of 0.008–0.035 Å, suggesting samples are monodisperse. Guinier analysis of the low-q regions indicates Rg values of 36.1 ± 0.2 Å averaged for the three concentrations with a Dmax value of 106 Å (for the apoprotein); no significant changes in Rg values were observed upon dilution. GASBOR22IQW was used for ab initio shape construction based on the best fit to the I(q) scattering data (Figure 6B,C). The apo form of the model (Figure 6B,C) has an overall shape consistent with the pentameric molluscan AChBPs determined by crystallography. The calculated Rg value of the crystal structure differs from the value based on the 1UV6 crystal structure (Rg = 33.4 Å). Additionally, a χ2 value of 5.1 is obtained by fitting the raw SAXS data to the ligand-bound crystal structure using CRYSOL (19).
FIGURE 6.
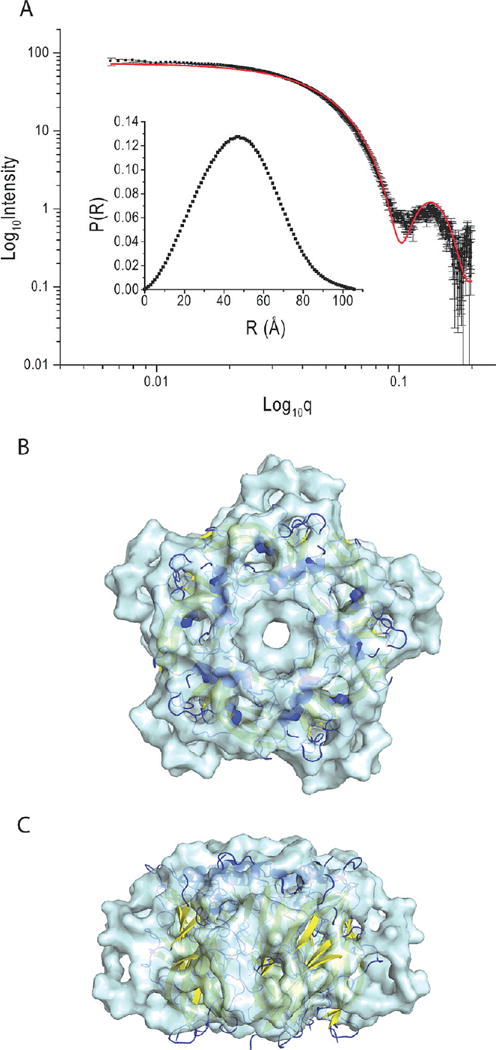
SAXS intensity data and P(r) distribution of the apo form of ct-AChBP. (A) SAXS intensity data for apo ct-AChBP (3.8 mg/mL), inset with the pairwise length distribution curve for the scattering particles. The solid red line is the computed SAXS intensity curve for the 1UV6 crystal structure. (B and C) SAXS structure determination of the ct-AChBP pentamer in the apo form from an axial (B) and lateral (C) view of the pentamer. The structure of Lymnaea AChBP 1UV6 (colored by secondary structure, cartoon representation) is superimposed. SAXS intensity data for other concentrations of ct-AChBP are shown in Figure S1 of the Supporting Information.
DISCUSSION
Understanding the structural details of binding of ACh and other agonists such as nicotine to the binding pocket of nAChRs has been greatly enhanced by the determination of the high-resolution crystal structure of AChBP, a protein thought to be secreted into the synapse by the glia of certain snails (25). Until now, the existence of AChBP in metazoans had been limited to the phylum Mollusca. One homologue of AChBP was found in the leech Haementeria ghilianii [included either in the mollusc phylum or in the annelid phylum, in different cladistic schemes (26)], but it appears to be β-subunit-like, suggesting that another protein (i.e., an α-subunit) would be required for functional assembly, as opposed to the homopentamer we and others have identified. The identification of AChBPs in other phyla provides clues toward understanding the evolutionary history of this protein (e.g., by providing an outgroup for the various mollusk AChBPs) and adds to what is known about variation in the sequence, structure, and function of the AChBP. The ct-AChBP sequence may also be useful for finding novel AChBPs in other annelid genomes, such as that of the earthworm, Lumbricus, or from other protostome phyla, such as Arthropoda. We show here that there are functional AChBP orthologs outside the phylum Mollusca, identified in a polychaete, phylum Annelida, with critical residues in the ligand-binding domain similar (but not identical) to previously characterized AChBPs.
The NMR-determined KD values for choline and nicotine are in good agreement with values obtained previously for a gliaderived AChBP from the mollusc L. stagnalis (25). The value for ACh binding was ~6-fold weaker, while the binding of α-bungarotoxin was found to be 42-fold weaker. This significant difference in affinity for α-bungarotoxin is not particularly surprising, given the fact that it exhibits a wide range of KD values for the three molluscan AChBPs as compared to agonists such as nicotine or ACh. In this context, it is notable that the Bulinus AChBP does not bind to α-bungarotoxin (9). The high affinity for nicotine relative to ACh in ct-AChBP is generally consistent with observations for molluscan AChBPs (27). The AChBPs and most nAChRs have an aromatic amino acid corresponding to the ct-AChBP amino acid at position 64 in the D loop. In the Capitella AChBP, this residue is substituted with isoleucine, which would not be expected to extend into the ligand-binding domain as far as large, aromatic residues such as Trp or Tyr. The observed KD for ACh of 28.7 μM provides some indication that this amino acid substitution, to date found uniquely in the polychaete AChBP, may be at least partly responsible for the decrease in the affinity of the protein for the physiologic ligand acetylcholine.
Structure determination has previously proven to be difficult for unliganded AChBPs (6); we present here the first low-resolution model of the apo form of AChBP. SAXS analysis of the apoprotein shows that ct-AChBP has an overall molecular envelope similar to those observed for molluscan AChBPs. However, comparison of the raw SAXS data of the ct-AChBP apoprotein (in solution) with the crystal structure of nicotine-bound ls-AChBP [CRYSOL χ2 value = 5.1 (19)] suggests that some structural differences do exist either between the two proteins or the two states (apo vs ligand-bound) of the proteins. These differences may be due to ligand-induced conformational changes, differences in the solution and crystalline conditions, or other factors (e.g., the presence of an attached oligosaccharide residue and/or the C-terminal tags). As SAXS analysis is limited by resolution, it remains to be determined with higher-resolution structural models of apo forms of AChBPs whether the differences between the apo and ligand-bound protein indicate global reorganization of the structure as previously suggested (3).
Physiologically, AChBPs have been proposed to be glial-secreted proteins that can affect synaptic transmission directly by buffering the ACh level of the synapses of snails (25). Interestingly, a recent study shows that most AChBP expression in the snail is nonsynaptic (7). Despite extensive structural studies of these binding proteins, their physiological role remains unclear. It will be of interest to discover why protostomes such as annelids and molluscs might rely upon AChBP as a stochiometric buffer of ACh, instead of (or in addition to) the enzyme acetylcholinesterase, which is widely found in metazoans and plays an important role in regulating synaptic transmission by reducing ACh levels.
Supplementary Material
Acknowledgments
We thank Christian Erxleben for critical reading of the manuscript and Changwon Lim for assistance with statistical calculations. SAXS data were collected at beamline X9 at the National Synchrotron Light Source at Brookhaven National Laboratory. Use of the X9 beamline was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886. We thank Lin Yang of the X9 beamline for data collection and beamline support.
Footnotes
Supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences. E.F.D.’s contribution was funded in whole with federal funds from the National Institutes of Health, National Institute of Environmental Health Sciences, under Delivery Order HHSN273200700046U to SRA International, Inc.
Abbreviations: nAChRs, nicotinic acetylcholine receptors; DSS, 4,4-dimethyl-4-silapentane-1-sulfonic acid; TMSP, trimethylsilylpropionate; ECD, extracellular domain; ACh, acetylcholine; IPD, ion pore domain; ls-AChBP, Lymnaea stagnalis AChBP; ac-AChBP, Aplysia californica AChBP; bt-AChBP, Bulinus truncatus AChBP; ct-AChBP, Capitella teleta AChBP; STD, saturation transfer difference; SAXS, small-angle X-ray scattering; EST, Expressed Sequence tag; KD, dissociation constant.
SUPPORTING INFORMATION AVAILABLE
Two supplemental figures, one showing SAXS data at two additional protein concentrations (Figure S1) and another showing additional mass spectrometry data (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 2.Le Novére N, Changeux JP. LGICdb: The ligand-gated ion channel database. Nucleic Acids Res. 2001;29:294–295. doi: 10.1093/nar/29.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gay EA, Yakel JL. Gating of nicotinic ACh receptors: New insights into structural transitions triggered by agonist binding that induce channel opening. J Physiol. 2007;584:727–733. doi: 10.1113/jphysiol.2007.142554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corringer PJ. Nicotinic receptors at the amino acid level. Annu Rev Pharmacol Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 5.Wilson G, Karlin A. Acetylcholine receptor channel structure in the resting, open, and desensitized states probed with the substituted-cysteine-accessibility method. Proc Natl Acad Sci USA. 2001;98:1241–1248. doi: 10.1073/pnas.031567798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 7.Banks G, Kemenes I, Schofield M, O’Shea M, Korneev SA. Acetylcholine binding protein of mollusks is unlikely to act as a regulator of cholinergic neurotransmission at neurite-neurite synaptic sites in vivo. FASEB J. 2009;23:3030–3036. doi: 10.1096/fj.08-117135. [DOI] [PubMed] [Google Scholar]
- 8.Hansen SB, Talley TT, Radic Z, Taylor P. Structural and ligand recognition characteristics of an acetylcholine-binding protein from Aplysia californica. J Biol Chem. 2004;279:24197–24202. doi: 10.1074/jbc.M402452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celie PH, Klaassen RV, van Rossum-Fikkert SE, van Elk R, van Nierop P, Smit AB, Sixma TK. Crystal structure of acetylcholine-binding protein from Bulinus truncatus reveals the conserved structural scaffold and sites of variation in nicotinic acetylcholine receptors. J Biol Chem. 2005;280:26457–26466. doi: 10.1074/jbc.M414476200. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Wang H, Cui Y, Zhang G, Zheng G, Liu S, Xie L, Zhang R. Mar Biotechnol. 2009;11:596–607. doi: 10.1007/s10126-008-9176-8. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Huang J, Sun J, Wang G, Li C, Xie L, Zhang RJ. A novel extrapallial fluid protein controls the morphology of nacre lamellae in the pearl oyster, Pinctada fucata. J Biol Chem. 2007;282:23253–23263. doi: 10.1074/jbc.M700001200. [DOI] [PubMed] [Google Scholar]
- 12.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 13.Lepre CA, Moore JM, Peng JW. Theory and applications of NMR-based screening in pharmaceutical research. Chem Rev. 2004;104:3641–3676. doi: 10.1021/cr030409h. [DOI] [PubMed] [Google Scholar]
- 14.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem, Int Ed. 2003;42:864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 15.Wang ZX. An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 1995;360:111–114. doi: 10.1016/0014-5793(95)00062-e. [DOI] [PubMed] [Google Scholar]
- 16.Seber GAF, Wild CH. Nonlinear Regression. Wiley; New York: 1989. [Google Scholar]
- 17.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 18.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svergun DI, Barberato C, Koch MH. CRYSOL: A Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 20.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2000;34:33–41. [Google Scholar]
- 21.Mukhtasimova N, Free C, Sine SM. Initial coupling of binding to gating mediated by conserved residues in the muscle nicotinic receptor. J Gen Physiol. 2005;126:23–39. doi: 10.1085/jgp.200509283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu CW, Wu FYH. Conformational transitions of cyclic adenosine monophosphate receptor protein of Escherichia coli. A temperature-jump study. Biochemistry. 1974;13:2573–2578. doi: 10.1021/bi00709a016. [DOI] [PubMed] [Google Scholar]
- 23.Gronenborn AM, Clore GM. Proton nuclear magnetic resonance studies on cyclic nucleotide binding to the Escherichia coli adenosine cyclic 3′,5′-phosphate receptor protein. Biochemistry. 1982;21:4040–4048. doi: 10.1021/bi00260a020. [DOI] [PubMed] [Google Scholar]
- 24.London RE, Perlman ME, Davis DG. Relaxation-Matrix Analysis of the Transferred Nuclear Overhauser Effect for Finite Exchange Rates. J Magn Reson. 1992;97:79–98. [Google Scholar]
- 25.Smit AB, Syed NI, Schaap D, van Minnen J, Klumperman J, Kits KS, Lodder H, van der Schors RC, van Elk R, Sorgedrager B, Brejc K, Sixma TK, Geraerts WP. Structure and function of AChBP, homologue of the ligand-binding domain of the nicotinic acetylcholine receptor. Nature. 2001;411:261–268. doi: 10.1038/35077000. [DOI] [PubMed] [Google Scholar]
- 26.Sixma TK, Smit AB. Acetylcholine binding protein (AChBP): A secreted glial protein that provides a high-resolution model for the extracellular domain of pentameric ligand-gated ion channels. Annu Rev Biophys Biomol Struct. 2003;32:311–314. doi: 10.1146/annurev.biophys.32.110601.142536. [DOI] [PubMed] [Google Scholar]
- 27.Celie PH, Kasheverov IE, Mordvintsev DY, Hogg RC, van Nierop P, van Elk R, van Rossum-Fikkert SE, Zhmak MN, Bertrand D, Tsetlin V, Sixma TK, Smit AB. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an α-conotoxin PnIA variant. Nat Struct Mol Biol. 2005;12:582–588. doi: 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


