FIGURE 3.
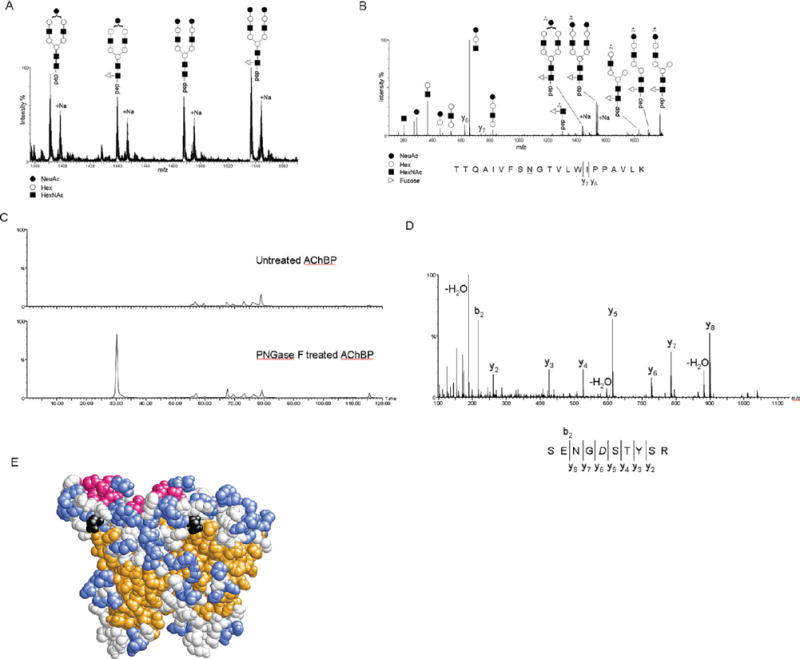
(A) Representation of the m/z range from 1380 to 1560 of an MS spectrum from a nanoLC-ESI-MS experiment and MS/MS analysis of the tryptic digest of ct-AChBP. Triply charged ions of the T17 peptide with Asn122 modified with the A1 (m/z 1390.34), A1F (m/z 1439.02), A2 (m/z 1487.34), and A2F (m/z 1536.07) types of complex glycans and the respective sodiated adducts. (B) Representation of the MS/MS spectrum of the A2F-modified T17 peptide from ct-AChBP. Generally, the CID-based fragmentation mechanism provides mostly information about the glycan structure. This can be seen by the extensive fragmentation of the glycan into its constituent mono-, di-, and trisaccharides, as well as pieces of the complex glycan remaining on the peptide. In this instance, MS/MS also provided sparse information about the peptide itself by the presence of the y6 and y7 ions. (C) Extracted ion chromatogram for the ion at m/z 558.2 for the untreated (top) and PNGase F-treated (bottom) ct-AChBP. The presence of this ion in only the PNGase F-treated ct-AChBP sample confirms Asn216 as a site of N-linked glycosylation. (D) MS/MS spectrum of the ion at m/z 558.2 that, via an extensive y-ion series, confirms the sequence of the peptide as SENGDSTYSR, where the italic D denotes the former site of glycosylation. (E) Space-filling model of the ct-AChBP homology model dimer. The predicted β-strands are colored orange, α-helices red, and loops and coils colored blue and white, respectively. The asparagine residue at position 122 is colored black.
