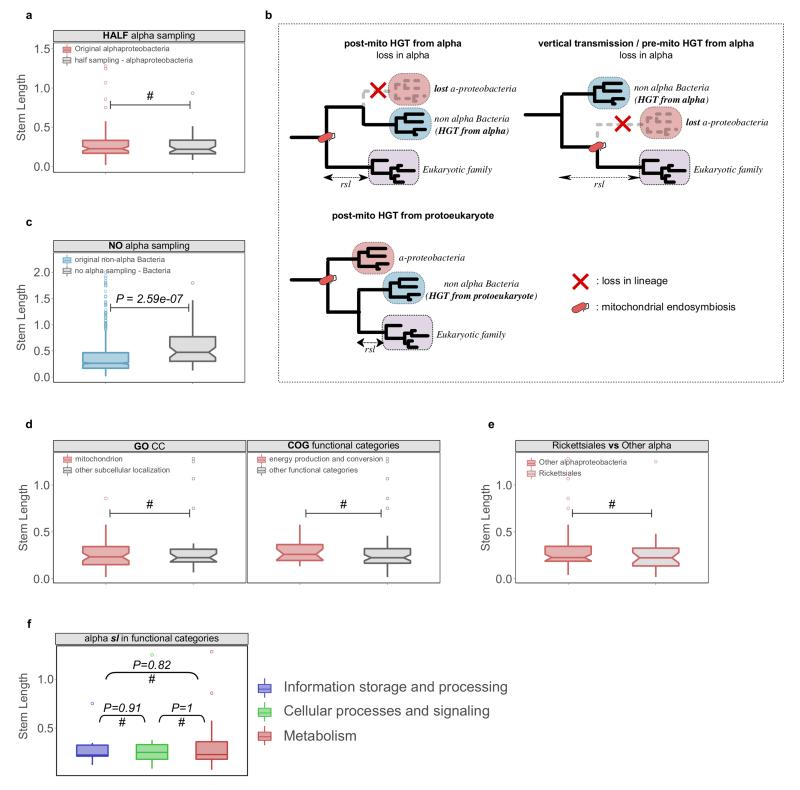
Extended Data Figure 6. Evaluation of alternative HGT scenarios and other potential biases.
a, The sampling effect was simulated by artificially removing part or all of the alpha-proteobacterial sequences in the final datasets. To simulate the potential bias caused by an enriched sampling of alpha-proteobacteria an artificial reduction of alpha-proteobacterial sequences to 50% was applied to the dataset (HALF alpha sampling). The reduction of alpha-proteobacterial sequences by 50% does not significantly change the inferred stem length within families of alpha-proteobacterial origin. b, Different scenarios of HGT to the proto-mitochondrion are unable to explain the observed signal in families mapped to non-alpha Bacteria. The transfer of a gene from alpha-proteobacteria to another bacterial lineage after mitochondrial endosymbiosis and its parallel loss from the lineage of the mitochondrial ancestor (“post-mito HGT from alpha”) would result in unchanged stem lengths. Loss of a gene from the alpha-proteobacterial sister clade would result in an increase of the inferred stem lengths (“vertical transmission / pre-mito HGT from alpha”). The transfer of a gene from the protoeukaryotic lineage to other bacterial clades would result in shorter stem lengths compared to the alpha-proteobacterial mappings (“post-mito HGT from protoeukaryote”). c, Upon total exclusion of alpha-proteobacterial sequences (NO alpha sampling), eukaryotic families map to other bacterial groups but with stem length higher than those observed typically. The same is observed when comparing the stem lengths of the families mapping to proteobacterial groups in the absence of alpha-proteobacteria, to those typically mapping to proteobacterial groups other than alpha-proteobacteria. d, Boxplots showing that there are no significant differences in the stem lengths between alpha-proteobacterial families with mitochondrial localization when compared to those with other subcellular localizations (left), or between families involved in energy related functions compared to those involved in other functional categories (right). e, Boxplot showing no significant difference between the distribution of stem lengths of families of Rickettsiales inferred origin and other alpha-proteobacteria. f, Alpha-proteobacterial families in different functional categories show no difference in stem lengths. In all the cases the distributions were compared using a two-sided Mann-Whitney U test. See also Supplementary Information sections 4-5.