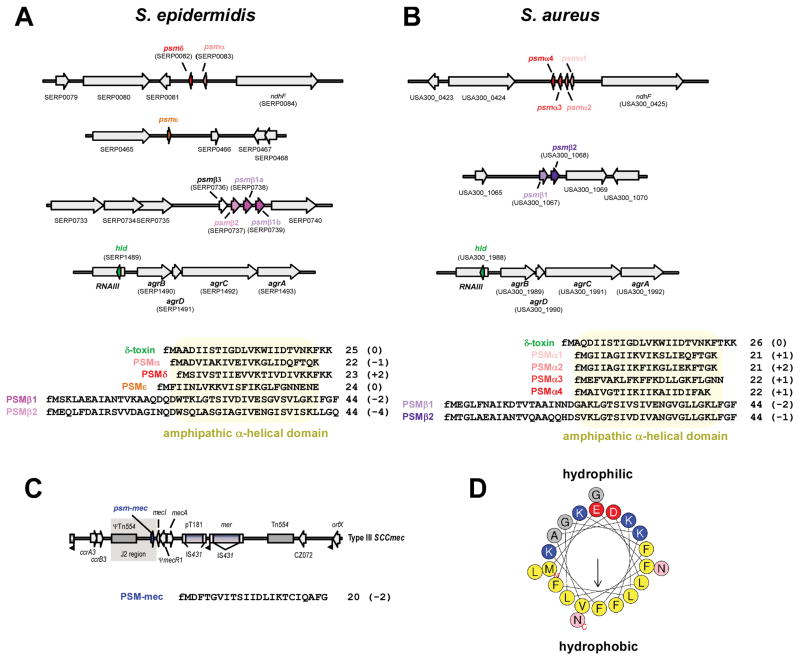
Figure 1. PSM genes, amino acid sequences, and structure.
(A,B) psm genes and amino acid sequences in S. epidermidis and S. aureus. Gene annotations are according to S. epidermidis strain RP62A and S. aureus strain USA300 FPR3757. All PSMs are secreted with an N-terminal N-formyl methionine (fM). Several α-type psm genes are not annotated in staphylococcal genomes owing to their short length. Note that the psmα and psmδ genes of S. epidermidis are located at a position in the genome corresponding to that of the S. aureus psmα operon, suggesting a common ancestor of these genes. The S. epidermidis psmβ operon contains a gene, psmβ3, whose gene product could not be detected in culture filtrates of S. epidermidis strains. Some S. epidermidis strains, such as RP62A, may contain two identical copies of the psmβ1 gene, resulting in higher PSMβ1 production than in strains that contain only one copy. The δ-toxin (sometimes called PSMγ), highly similar between S. epidermidis and S. aureus, is encoded by the gene “hld” (for “hemolysin delta”), located within RNAIII in the Agr system. (C) Location of the psm-mec gene in SCCmec elements. The psm-mec gene is found in SCCmec elements of types II, III, and VIII, in the J2 region next to the class A mec gene complex (with the core genes of the SCCmec element in the order IS431-mecA-mecR-mecI), which is characteristic for these SCCmec types. Type III is shown here as example. (D) α-helical wheel presentation of PSMα3, showing the extreme amphipathy that is characteristic of PSMs, with hydrophobic and hydrophilic amino acids found on opposite sides of the α-helix. (A–C), numbers behind amino acid sequences show peptide length and charge (in parentheses).