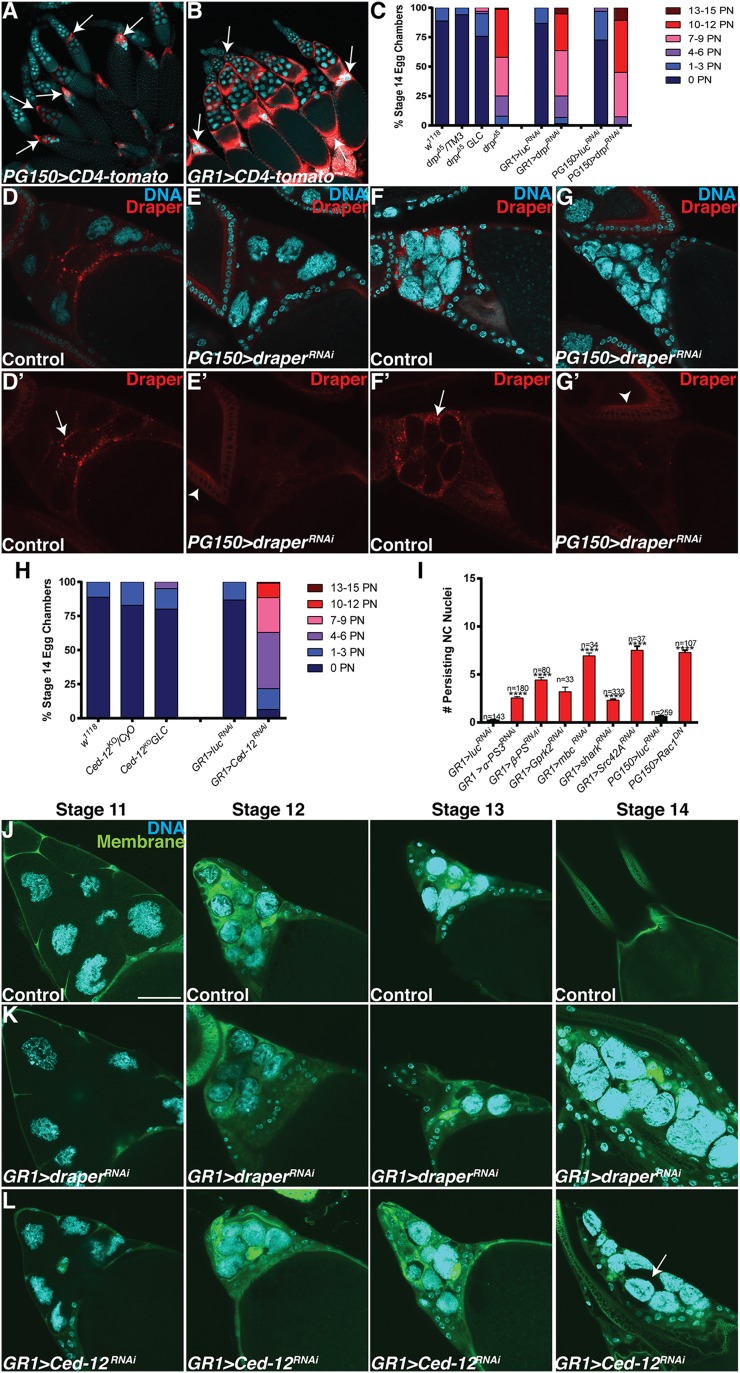
Fig. S1.
FC GAL4 drivers and analysis of draper, Ced-12, and other engulfment genes. (A and B) FC GAL4 driver expression patterns. Egg chambers express the membrane-tethered protein UAS-CD4-tomato (red) specifically in the FCs and are stained with DAPI (cyan) to label DNA. (A) PG150-GAL4 drives expression of UAS-CD4-tomato (PG150-GAL4/+;UAS-CD4-tomato/+) specifically in the stretch FCs beginning in stage 10 through stage 14 (arrows). (B) GR1-GAL4 drives expression of UAS-CD4-tomato (UAS-CD4-tomato/+; GR1-GAL4/+) in all FCs (arrows) beginning in stage 3. (C) Alternative quantification of data presented in Fig. 1Q. The number of persisting nuclei (PN) per stage 14 egg chamber was categorized into bins of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, and 13–15 PN, and the percentage of stage 14 egg chambers in each bin was calculated. (D–G’) The enrichment of Draper is specific to the FCs. Late stage egg chambers expressing mCD8-GFP (control) or draperRNAi specifically in the stretch FCs were stained together with α-Draper antibody (red, arrows) and DAPI (cyan). (D and D’) Control (PG150-GAL4/+; UAS-mCD8-GFP/+) stage 11 egg chamber shows enriched Draper along membranes that surround the NCs. (E and E’) draper is knocked down specifically in the stretch FCs of a stage 11 egg chamber (PG150-GAL4/+; UAS-draperRNAi/+), and the enrichment of Draper is absent. Basal levels of Draper staining are apparent in an earlier stage egg chamber (arrowhead). (F and F’) Control (PG150-GAL4/+; UAS-mCD8-GFP/+) stage 13 egg chamber shows enriched Draper along membranes that surround the NCs. (G and G’) draper is knocked down specifically in the stretch FCs of a stage 13 egg chamber (PG150-GAL4/+; UAS-draperRNAi/+), and the enrichment of Draper is absent. Basal levels of Draper staining are apparent in an earlier stage egg chamber (arrowhead). (H) Alternative quantification of data presented in Fig. 1R. The number of persisting nuclei (PN) per stage 14 egg chamber was categorized into bins of 0 PN, 1–3 PN, 4–6 PN, 7–9 PN, 10–12 PN, and 13–15 PN, and the percentage of stage 14 egg chambers in each bin was calculated. (I) Quantification of persisting NC nuclei in engulfment gene knockdowns analyzed in the candidate gene screen (Table S1). Data presented are mean ± SEM. ****P ≤ 0.0001. (J–L) Visualization of stretch follicle membranes with mDC8-GFP. (Scale bar, 50 μm.) (J) Stage 11–14 egg chambers from GR1-GAL4 UAS-mCD8-GFP/TM3 (control) flies express membrane–GFP in all FCs (green) and are stained with DAPI (cyan). The stretch FC membranes surround the NCs during late oogenesis. (K) Most FCs from UAS-draperRNAi/+; GR1-GAL4 UAS-mCD8-GFP/+ egg chambers in stages 10–14 of oogenesis appear to surround the NCs. (L) Most FCs from GR1-GAL4 UAS-mCD8-GFP/UAS-Ced-12RNAi egg chambers in stages 11–14 of oogenesis appear to surround the NCs. Occasionally some NCs do not appear to be surrounded by the FCs (arrow).