Fig. S5.
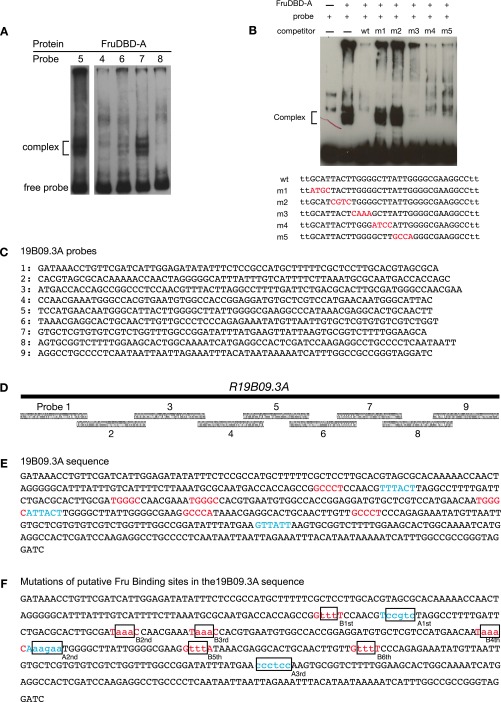
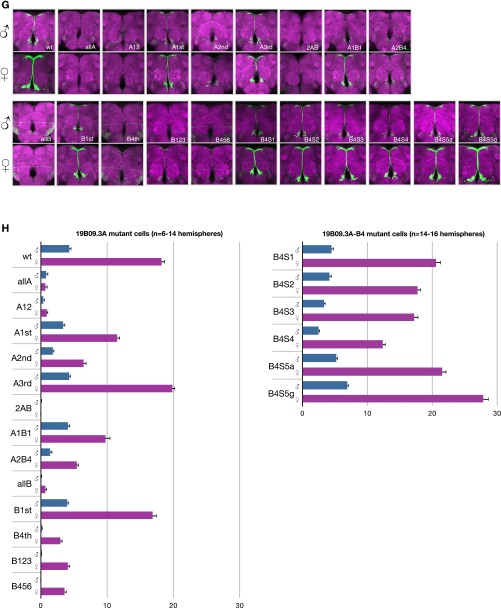
(Related to Fig. 3.) R19B09.3A Fru binding and mutations. (A) Fru-DBD-A shows binding to R19B09.3A probes #5 and #7 but weak or no binding to other probes by EMSA. There appear to be multiple bands for the DNA–protein complex (as indicated), most likely a result of the partial degradation of the fusion protein. Data are shown for five of the nine tested probes. (B) Fru-DBD-A binding to a shorter probe derived from probe #5 was assayed in competition with probes containing tiled substitutions. The labeled probe consisted of the wild-type (wt) sequence in the lower part of the figure. The mutant sequences used as competitors are listed below with the substituted nucleotides indicated in red. The wild-type competitor effectively competed with probe binding, but m1 and m2 mutations prevented competition, suggesting these regions are necessary for Fru-DBD-A binding. (C) Sequences of probes tiling R19B09.3A used for EMSA. (D) Diagram of probes from C tiling across R19B09.3A. (E) Summary of the potential Fru-DBD-A and Fru-DBD-B binding sites identified by EMSA in R19B09.3A. The binding sites for Fru-DBD-A are indicated in red and the binding sites for Fru-DBD-B are indicated in blue. (F) Mutations made for the Fru binding sites in the R19B09.3A sequence. The positions of binding sites for Fru-DBD-A are indicated in red and the positions of binding sites for Fru-DBD-B are indicated in blue. The mutated nucleotides at each binding site are indicated in lowercase. (G) Mutations were made in various combinations to assay effects in vivo based on EMSA results, as labeled. For example, “allA” included mutations at all Fru-DBD-A sites, whereas “A12” mutated Fru-DBD-A sites 1 and 2. “B4S1” through “B4S5g” were single base pair changes across the Fru-DBD-B binding site 4, with “B4S5a” being a mutation of C to A and “B4S5g” being a mutation of C to G. Images were captured at 20× magnification. (H) Cell counts from G. Error bars indicate SEM.