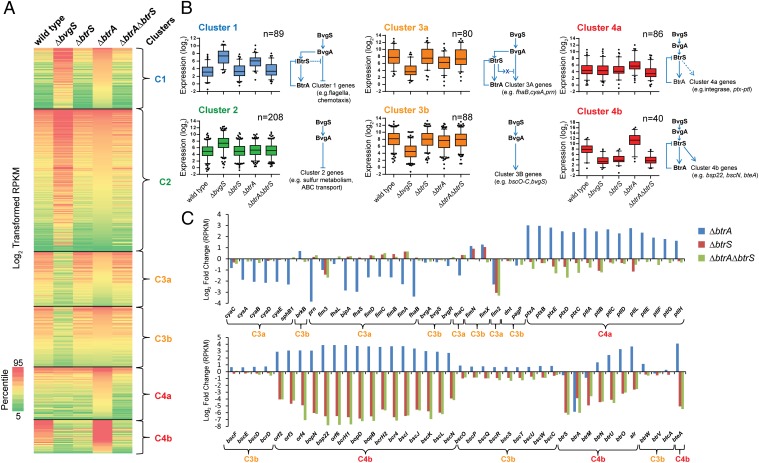
Fig. 4.
BtrA, BtrS, and BvgS regulatory networks identified using RNA-seq. (A) Heat map of log2 transformed reads per kilobase of transcript per million mapped reads (RPKM) values showing expression levels of specific transcripts (rows) in samples from WT RB50 or ∆bvgS, ∆btrS, ∆btrA, or ∆btrA∆btrS derivatives (columns). Genes with at least plus or minus threefold change in expression relative to WT were included in the analysis. Nonhierarchical clustering of RPKM values revealed four coexpressed gene clusters (C1–C4). Map colors indicate the percentile score of expression level. RNA-seq alignment statistics, datasets, and comparative analyses are listed in Table S4 and Dataset S1. (B) Box-and-whisker plots showing the distribution of transcripts within each gene cluster. Boxes depict data between the 25th and 75th percentiles, with horizontal lines within boxes representing the median value. Whiskers show 5th and 95th percentiles, with outliers as dots. The projected regulatory network for each cluster is drawn next to the respective plot. Arrowheads indicate transcriptional activation, and bars indicate repression. For cluster 3a, “X” designates a predicted BtrS-activated repressor that acts on a subset of BvgAS-regulated genes. (C) Log2-fold changes in expression levels of selected genes in deletion mutants compared with WT RB50. (Top) Genes are ordered by relative position in the RB50 genome. (Bottom) T3SS genes ordered by relative position. Curly brackets designate coexpression clusters.