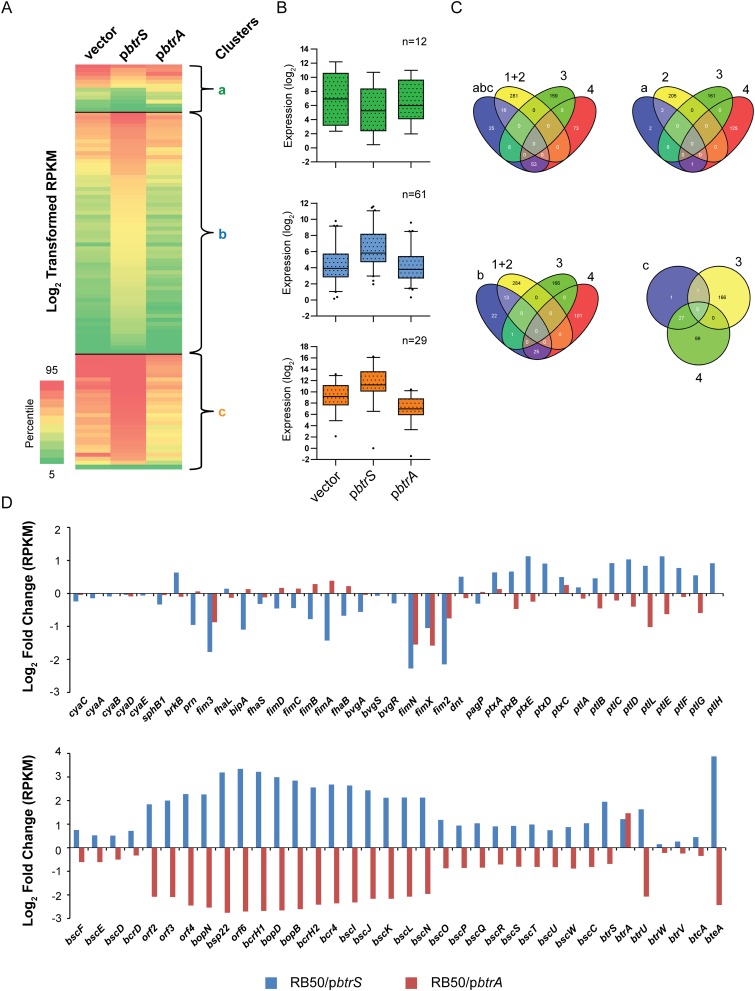
Fig. S5.
Overexpression of btrA or btrS corroborate regulatory networks predicted by RNA-seq analysis of deletion mutants. (A) Heat map of log2-transformed RPKM values showing expression levels of specific transcripts (rows) in samples from WT RB50/vector, RB50/pbtrS, or RB50/pbtrA (columns). Genes with at least plus or minus threefold perturbation in expression relative to vector-only were analyzed. Nonhierarchical clustering of RPKM values was performed to derive coexpressed gene clusters (clusters a–c). Colors on map indicate percentile relative expression levels. (B) Box-and-whisker plots showing distribution of transcripts within each cluster. Boxes depict data between the 25th and 75th percentiles, with horizontal lines within boxes representing the median value. Whiskers show 5th and 95th percentiles, with outliers as dots. (C) Venn diagrams showing the number of genes that overlap between null-mutants (Fig. 4) and after overexpression of btrA and btrS. Areas within Venn diagrams are not scalable. The Venn diagrams were created using Venny (bioinfogp.cnb.csic.es/tools/venny/). Of the 90 genes that displayed significantly increased transcription after overexpression of BtrS, 29 were down-regulated by BtrA overexpression and all were categorized as BtrS-activated and BtrA-repressed in the analysis in Fig. 4 (clusters 4a and 4b). Of the 80 loci with decreased expression in the ∆btrA strain (cluster 3a), none were further induced in RB50 by overexpressing BtrA. Although results from deletion vs. overexpression strains are congruent, complete correspondence is not expected. If the levels of BtrA in RB50 exceed the threshold for maximal activation or repression of a given gene, for example, overexpression will have no effect as observed with cluster 3a genes. See also Dataset S1. (D) Log2-fold change in gene expression of known virulence-related genes compared with WT.