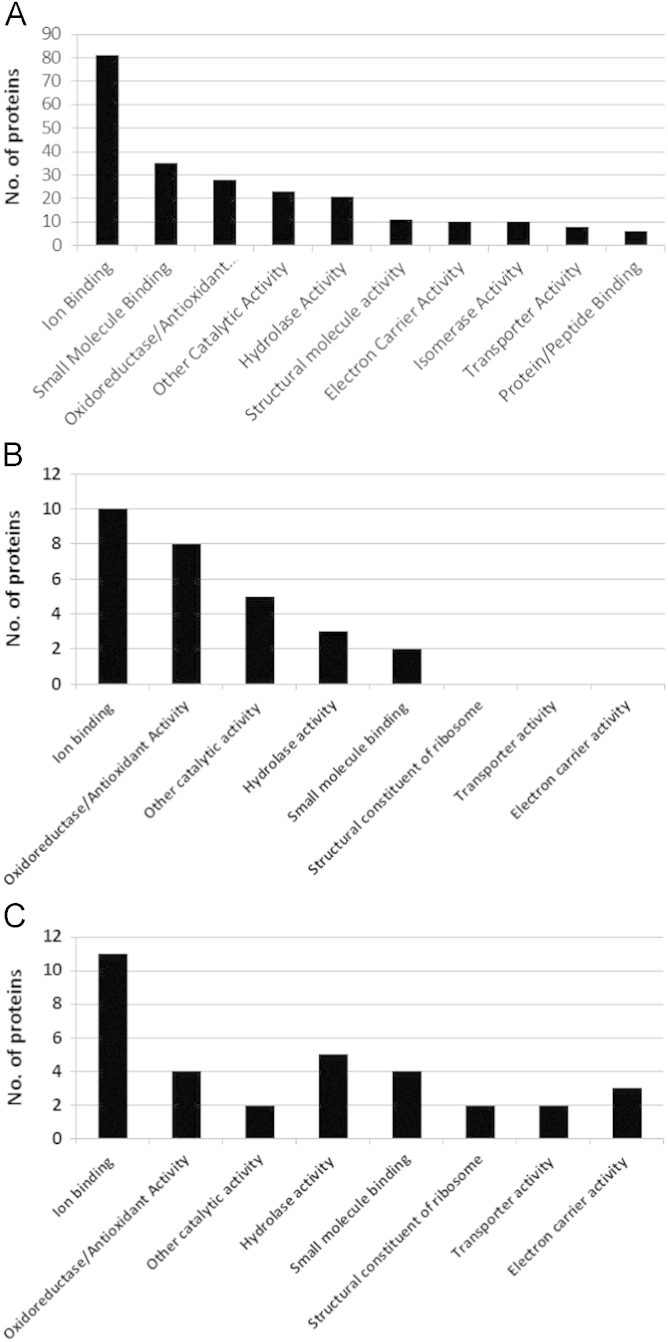
Fig. 3.
Functional classifications (according to GO terms) of the trichome-enriched sample proteins identified from Mascot searches against the (A) Uniprot viridiplantae׳ database, (B) ׳York Artemis contigs׳ database for protein identifications with higher abundance in trichome-enriched sample material, and (C) ׳York Artemis contigs׳ database for protein identifications with lower abundance in trichome-enriched sample material. Mascot search results were submitted to Percolator with an ׳expect cut-off׳ threshold of 0.05 and filtered using a minimum number of significant sequences of 2. Adapted from [1].