Figure 6.
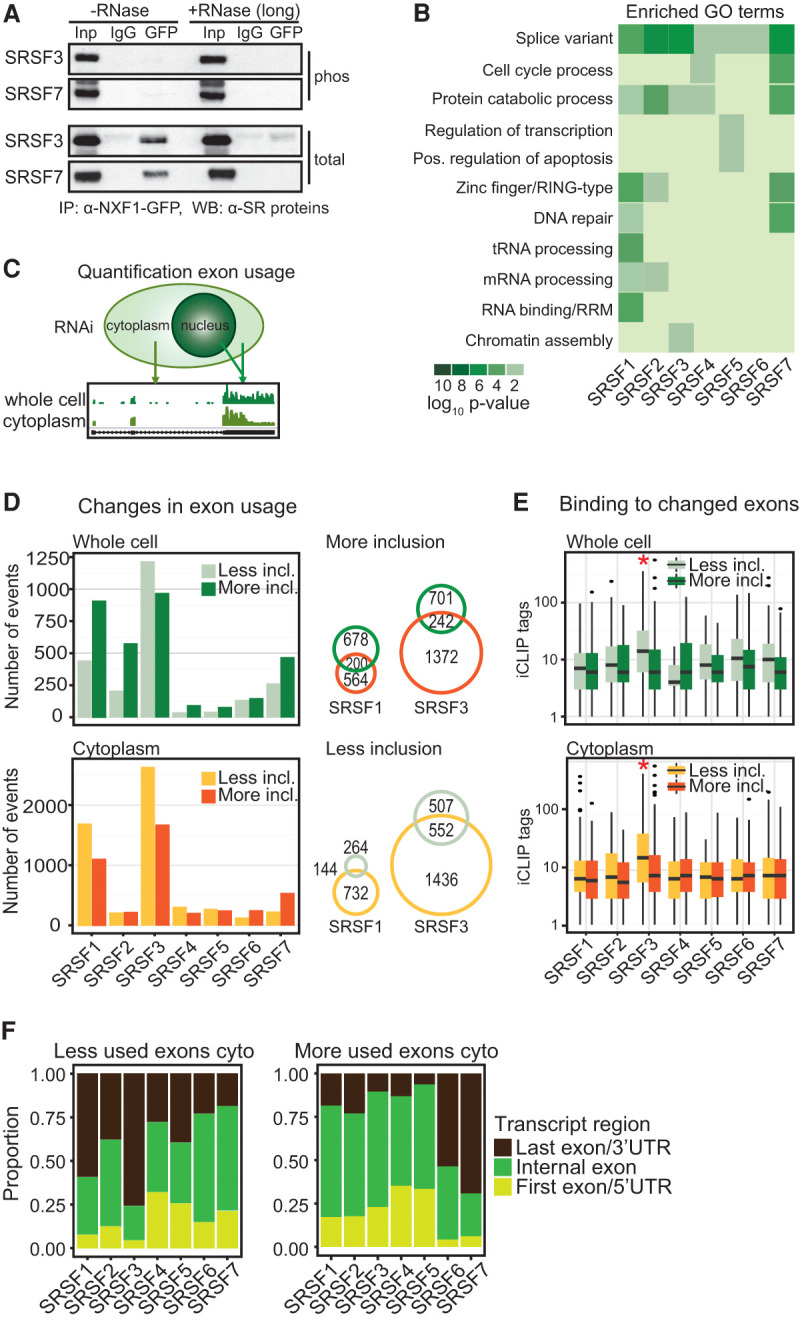
SR proteins link alternative splicing to mRNA export. (A) Co-IP of NXF1-GFP with or without RNase A (long) using α-GFP. Phosphorylated SRSF3 and SRSF7 were detected with phosphorylation-dependent mAb104 (top panel) and phosphorylation-independent SR protein-specific (bottom panel) antibodies. (B) Gene ontology (GO) term analysis of export targets with significant cross-links of their cognate SR proteins. (C) Quantification of exon usage from cytoplasmic and whole-cell RNA-seq after SR protein depletion. (D, left panel) Changes in exon usage in whole-cell and cytoplasmic RNA samples were calculated using DEX-seq (Anders et al. 2012). P < 0.01. Numbers of changed exons after SR protein knockdown were separated in less inclusion and more inclusion. (Right panel) Venn diagrams showing coregulated splicing events in total and cytoplasmic samples after SRSF1 and SRSF3 depletion. (E) Number of significant cross-link events in changed exons in whole-cell and cytoplasmic RNA. (F) Proportion of exons that changed after SR protein knockdowns separated into first exons (light green), internal exons (dark green), and last exons (dark brown).
