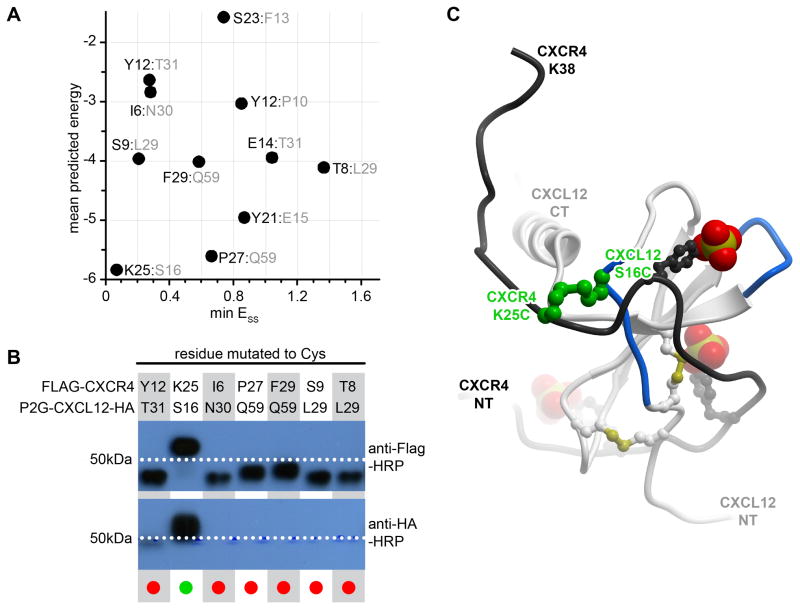
Figure 4.
Identification of the first CRS1 crosslink between CXCR4 and CXCL12 from the CXCR4:CXCL12 NMR structure (PDB 2k05). (A) 11 crosslinks were designed in silico and ranked according to the overall predicted energy (vertical) and disulfide bond energy (horizontal). (B) Following co-expression of the mutant pairs in insect cells, the best ranking pair, CXCR4(K25C):CXCL12(S16C) demonstrated efficient crosslinking in Western blotting as evidenced by the double (anti-receptor and anti-chemokine) staining and the molecular weight shift of the material in the respective lane. (C) The identified crosslink (green) is shown in the context of the NMR structure from which it was generated: CXCL12 is in white ribbon and the N-terminal peptide of CXCR4 is in black ribbon. The sulfate groups on CXCR4 sulfotyrosines sTyr7, sTyr12, and sTyr21 are shown as CPK. Panel (B) is adapted from (Kufareva et al., 2014).