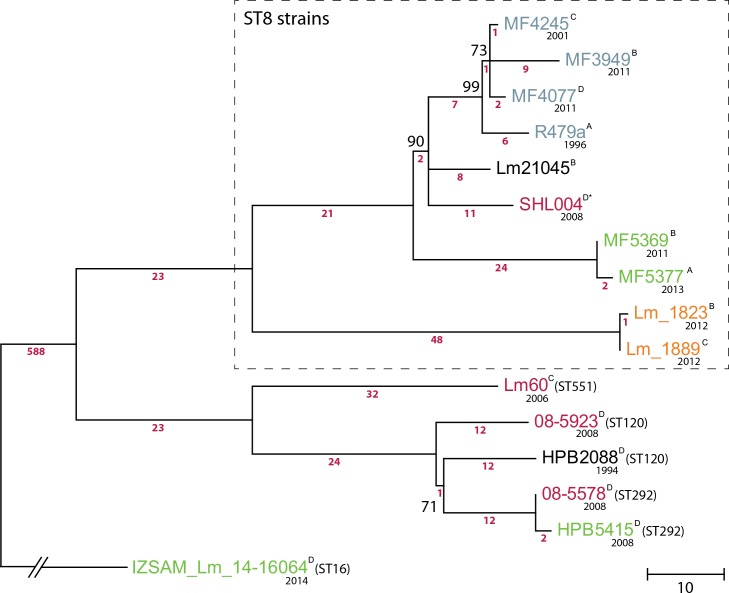
Fig 1. Phylogeny for the L. monocytogenes CC8 isolates based on whole-genome SNP analysis.
Maximum Parsimony tree based on 872 non-homoplasic chromosomal SNPs. The tree was rooted on L. monocytogenes IZSAM_Lm_14–16064. Bootstrap support is shown for nodes with <100% bootstrap support. Branch lengths are in the units of the number of changes over the whole sequence (except for the branch for IZSAM_Lm_14–16064 which has been shortened for clarity), and the number of contributing SNPs is indicated below each branch (numbers in pink). For details on the identity of the indicated SNPs differentiating the strains, see S1 and S2 Tables. The ST8 lineage is indicated by a box with dashed line, for the other strains the ST is indicated in parenthesis after strain names. The origin of strains is indicated by the colour of the strain name: Blue; salmon processing facility, green; poultry processing facility or meat, orange; cheese, pink; human clinical strain. Superscripts after strain names indicate the variant of the hsdRMS locus found in each strain (see text, Fig 2 and Table 3 for details). The year of isolation (when known) is noted below the name of each isolate.