Abstract
Importance
The intestinal microbiome plays a critical role in infant development, and delivery mode and feeding method (breastmilk vs. formula) are determinants of its composition. However, the importance of delivery mode beyond the first days of life is unknown, and studies of associations between infant feeding and microbiome composition have been generally limited to comparisons between exclusively breastfed and formula fed infants, with little consideration given to combination feeding of both breastmilk and formula.
Objectives
To examine the relative effects of delivery mode and feeding method on infant intestinal microbiome composition at approximately six weeks of life.
Design, Setting and Participants
Prospective observational study of 102 infants followed as part of a US pregnancy cohort study.
Exposures
Delivery mode was abstracted from delivery medical records and feeding method prior to the time of stool collection was ascertained through detailed questionnaires.
Main Outcomes and Measures
Stool microbiome composition was characterized using next-generation sequencing of the 16S rRNA gene.
Results
We identified independent associations between microbial community composition and both delivery mode and feeding method. Differences in microbial community composition between vaginally and infants delivered by Cesarean section were equivalent to or significantly larger than those between feeding groups. Bacterial communities associated with combination feeding were more similar to those associated with exclusive formula feeding than exclusive breastfeeding. We identified individual bacterial genera that were differentially abundant between delivery mode and feeding groups.
Conclusions and Relevance
The infant intestinal microbiome at approximately six weeks of age is significantly associated with both delivery mode and feeding method, and the supplementation of breastmilk feeding with formula is associated with a microbiome composition that resembles that of infants who are exclusively formula fed. These results may inform feeding choices and shed light on the mechanisms behind the lifelong health consequences of delivery and infant feeding modalities.
Keywords: microbiome, early childhood, breastfeeding, cesarean section
Introduction
Following birth and the initiation of feeding, the human gastrointestinal tract is colonized by a large diversity of bacterial life. An emerging body of literature in adults has begun to establish clear relationships between gut microbiome composition and a wide range of health outcomes 1–6. In contrast, comparatively little is known about the gut microbiome in infants and children, the exposures that shape it, and its lifelong health impacts 7. Although limited in their size and scope, a number of studies have established relationships between intestinal microbiome profiles in infants, delivery mode and/or breastmilk exposure 8–15. These factors both have long-term health consequences. Cesarean delivery has been associated with an increased risk of obesity, asthma, celiac disease and type 1 diabetes 16–19, whereas breastfeeding has been related to decreased risks of asthma, obesity, infection, metabolic syndrome, and diabetes, among other illnesses compared with formula feeding (reviewed in 20). The underlying mechanisms are not well understood, but there is growing evidence linking exposure to microflora that is present during vaginal delivery with the patterns of the microbiome that become established in infants 21. In addition, following delivery, the feeding of human milk primes and matures the infant gastrointestinal system, and is believed to promote a unique microbial colonization profile that has yet to be clearly defined in healthy populations 22. The acquisition of specific microbes in succession, as the core microbiome of the gut is created, may be permanently affected by exposure to maternal vaginal microflora and/or to breastmilk and could represent a key mechanism underlying differences in immune development that influence lifelong disease risk.
The contribution of bacteria through vaginal delivery followed by exclusive breast feeding promotes specific microbial profiles that facilitate optimal nutrient metabolism and early systemic immune training 23. The potential short and long term effects of perturbations of the gut microbiome of infancy as influenced by operative delivery or formula feeding are beginning to be examined. The contribution of mode of delivery to the infant microbiome has been evaluated 13,15,24. However, no study has examined the effects of delivery mode and breastfeeding following adjustment for the other, and there is little data on the effects of combination feeding—feeding breastmilk and formula together. Determining the associations between mode of delivery and breastmilk versus formula feeding and microbiome development in infants is critical to informing delivery and feeding decisions or interventions to alter the microbiome for improved health. Our objective was to evaluate the relative effects of delivery and feeding modes on the composition of the intestinal microbiota at approximately six weeks of age in 102 infants from a US pregnancy cohort study. The observed differences due to delivery and feeding modes highlight their importance in shaping the early intestinal microbiome and point to possible explanations for some of the risks and benefits associated with infant delivery and feeding practices.
Methods
Ethical approval, informed consent and privacy
Institutional review board approval was obtained from the Center for the Protection of Human Subjects at Dartmouth with yearly renewal of approval, and subjects provided written informed consent to participate and to permit their children to participate.
The New Hampshire Birth Cohort Study
Pregnant women ages 18–45 were recruited from prenatal clinics beginning at approximately 24 to 28 weeks gestation as described previously 25,26. We performed microbiome characterizations of stool samples collected at approximately six weeks of age from full term infants (>37 weeks gestational age at delivery, and appropriate growth for gestational age). Six weeks was chosen as it is likely that exclusive breastmilk or formula feeding would be well established at this age, and six weeks corresponded to routine maternal postpartum visits, a time that allowed for optimal sample collection with minimal particpant burden. We evaluated infant diet from birth until the time of stool collection by telephone questionnaires that included questions regarding the duration of breastfeeding and the timing of formula introduction, if any. Infants who were fed breastmilk and who had never been given formula prior to the time of stool collection were given the status of exclusive breastmilk feeding; infants who had not been breastfed and who had been fed formula only prior to their stool collection were assigned the status exclusively formula-fed; and infants who had received both breastmilk and formula prior to their stool collection were identified as having a diet of both breastmilk and formula. When possible, we confirmed exclusive breastmilk and exclusive formula feeding status using a feeding diary kept by subjects’ mothers during the 48-hour period prior to stool collection.
Delivery mode (Cesarean vs. vaginal delivery) was abstracted from maternal delivery records. Data about infant exposures to medication were derived from questions asked during the telephone questionnaires described above. Mothers were asked whether their infant had received a prescription medication in the first four months of life. A free text field was used to record the medication name. If the exact name could not be recalled, as much detail as could be recalled was recorded. Topical medications including those given for conjunctivitis and antifungals such as those given for thrush were not considered. Because antibiotic exposure has both been shown to influence the intestinal microbiome 7 we excluded infants who had received a prescription antibiotic.
Sample collection, DNA extraction and sequencing
Study participants provided infant stool samples collected at regularly scheduled maternal postnatal follow-up visits (six weeks post partum). Stool was aliquoted in sterile tubes and frozen at −80°C within 24 hours of receipt. Samples were thawed and DNA was extracted using the Zymo DNA extraction kit (Zymo Research). Quantity and purity of the DNA were determined by OD260/280 nanodrop measurement. Reliability and stability of these methods were described by Wu et al 27. Illumina tag sequencing of the 16S rRNA gene V4–V5 hypervariable region was performed at the Marine Biological Laboratories (MBL) in Woods Hole, MA using established methods 28,29. Details of sequencing methods, quality control and filtering, and statistical modeling are presented in the Supplement (see eMethods).
Results
Study subject characteristics and variability and diversity of the early neonatal microbiome
We evaluated the relationships between the composition of the six-week intestinal microbiome and both delivery mode and feeding method in 102 full term, appropriately grown infants enrolled in the New Hampshire Birth Cohort Study. Delivery medical records, telephone surveys and feeding diaries were used to assess study participant characteristics including delivery mode and feeding method at the time of stool sample collection (table 1). We found no significant relationship between delivery mode and feeding method (see eTable in the Supplement; Fisher exact test: p=0.66).
Table 1.
Subject characteristics (n=102)
| Variable | Mean (range) or % |
|---|---|
| Gestational age (weeks) | 39.7 (37.1–41.9) |
| Delivery mode | |
| Vaginal | 69% |
| Cesarean section | 31% |
| Infant sex | |
| Male | 54% |
| Female | 46% |
| Infant birth weight (g) | 3530 (2700–4710) |
| Feeding at six weeks | |
| Exclusively breastfed | 69% |
| Combination feeding | 25% |
| Exclusively formula fed | 6% |
| Age at formula introduction among combination fed subjects (weeks) | 3.1 (0.1, 8.7) |
We sequenced the V4–V5 regions of the bacterial 16S rDNA to characterize the microbial communities present in a stool sample from each study subject at six weeks of age. Sequencing yielded a total of 14,362,739 (mean: 140,811, range: 27,897 – 260,579) bacterial DNA reads, of which, 8,210,402 (mean: 80,494, range: 12,244 – 178,802) passed quality filters (see eMethods in the Supplement). These were assigned to 241 bacterial genera. Over 90% of reads were represented by 10 genera (table 2). Stool samples were dominated by Bacteroides and Bifidobacterium comprising half of sequence reads, with Streptococcus, Clostridium, Enterococcus, Blautia, Veillonella, Lactobacillus, Staphylococcus, Planococcus, and others representing the remainder (table 2).
Table 2.
Relative abundance of the 10 most abundant bacterial genera identified, for all subjects overall and for individual delivery mode and feeding groups.
| Genus | Overall (n=102) (%) | Vaginally delivered (n=70) (%) | Delivered by C-section (n=32) (%) | Exclusively breastfed (n=70) (%) | Combination fed (n=26) (%) | Exclusively formula fed (n=6) (%) |
|---|---|---|---|---|---|---|
| Bacteroides | 26.4 | 34.6 | 20.7 | 27.9 | 22.1 | 28.8 |
| Bifidobacterium | 22.5 | 23.3 | 17.4 | 25.5 | 16.8 | 11.4 |
| Streptococcus | 13.8 | 12.1 | 14 | 11.7 | 18.7 | 16.9 |
| Clostridium | 7.9 | 5.1 | 8.8 | 6.8 | 11.9 | 2.4 |
| Enterococcus | 5.7 | 4.3 | 8.7 | 4.8 | 6.1 | 14.6 |
| Blautia | 3.6 | 2.7 | 5.5 | 1.8 | 7.1 | 9.4 |
| Veillonella | 3.4 | 3.6 | 4.6 | 3.5 | 3.2 | 2.9 |
| Lactobacillus | 3 | 2.5 | 4.2 | 3.4 | 2.8 | 0 |
| Staphylococcus | 2.6 | 1.6 | 3.4 | 3.3 | 1.2 | 0.1 |
| Planococcus | 2 | 1.4 | 2.9 | 1.5 | 3.3 | 2.6 |
| Other genera | 9.1 | 8.8 | 9.8 | 9.8 | 6.8 | 10.9 |
Relationships between delivery mode, feeding method and microbial community composition
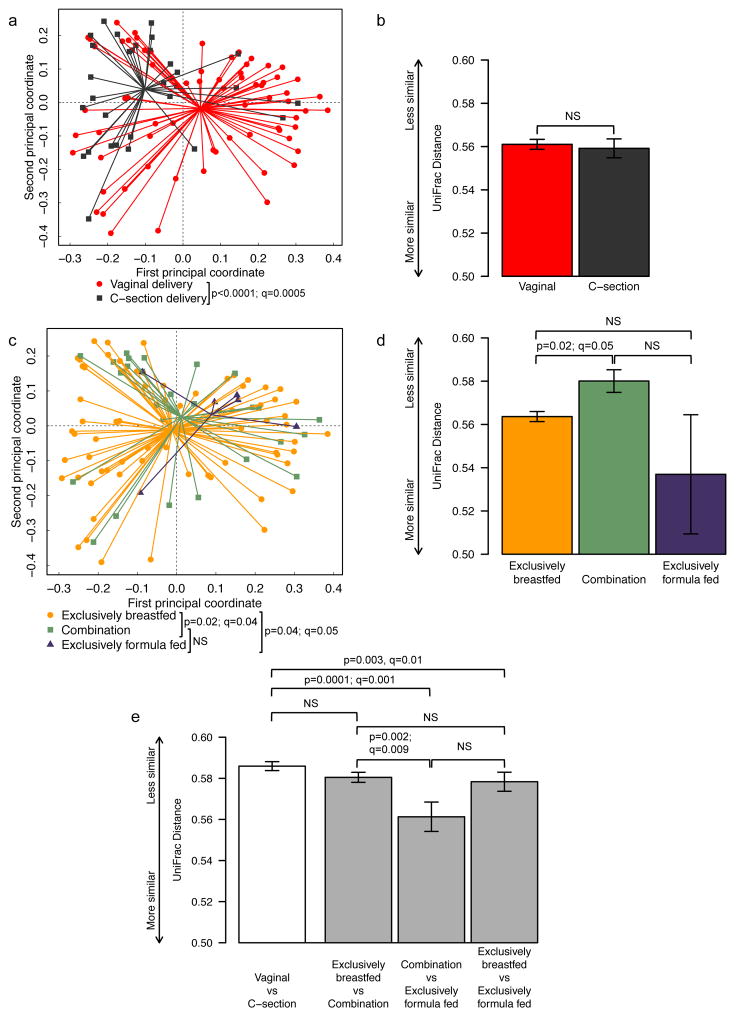
Overall stool microbiome community composition was characterized using generalized UniFrac analysis 30. Controlling for the effects of feeding method, delivery mode was strongly associated with infant gut microbiome composition (p<0.0001, q=0.0005) (figure 1a). Likewise, controlling for the effects of delivery mode, the overall association between feeding method and stool microbiome community composition was also statistically significant (p=0.01, q=0.02) (figure 1c). In pairwise comparisons of the three feeding methods, exclusive breastfeeding was associated with a microbiome community distinct from that of infants who were either exclusively formula-fed (p=0.04, q=0.05) or fed a combination of breastmilk and formula prior to stool collection (p=0.02, q=0.04). There was no statistically significant difference between infants fed a combination of breastmilk and formula and those fed exclusively formula in terms of microbiome composition. There was no significant interaction observed between delivery mode and feeding method (p=0.49). The lack of an interaction between delivery mode and feeding method remained even after combining exclusively formula fed and combination fed infants into single group (p=0.53).
Fig. 1. Comparison of microbial community composition between delivery mode and feeding method groups.
Principal coordinate plots (a, c) and mean pairwise UniFrac distances (b, d, e) within groups for delivery mode (a, b) and feeding method (c, d) and between groups (e). UniFrac is a distance metric used for comparing biological communities that incorporates information on the phylogenetic relatedness of community members. For (a) and (c), individual subjects are represented by points marked according to delivery mode (a) or feeding method (c) and are plotted on the first two principal coordinates with ADONIS p-values indicated. Lines are drawn from each point to its group centroid. For (b), (d) and (e), bar height is proportional to mean pairwise UniFrac distance within (b) and (d) or between (e) groups with error bars indicating standard error of the mean. In (e), white bars indicate delivery mode groups while gray bars indicate feeding method groups. Throughout, Q-values indicate significance of differences after adjusting for multiple comparisons by controlling the false discovery rate for selected comparisons.
We calculated within- and between-group average UniFrac distances in order to assess the group-specific phylogenetic diversity of the microbial communities we observed in our subjects. Within-group distances between infants within specific delivery mode and feeding method groups revealed similar average phylogenetic distances among infants who were born vaginally compared with those born by Cesarean section (figure 1b). The greatest within-group average pairwise phylogenetic distance was observed among those infants who were fed breastmilk supplemented with formula; pairs within this group were on average significantly less similar than pairs within the exclusively breastfed group (p=0.02, p=0.04), while other comparisons did not reach statistical significance (figure 1d).
Between-group pairwise UniFrac distances were, on average, as large or larger between vaginal and Cesarean section-delivered infants as they were between infants from different feeding groups (figure 1e). For feeding method, average bacterial community phylogenetic distance was greatest between infants who were exclusively breastfed as compared to those exclusively formula fed and between infants who were exclusively breastfed as compared to those who were fed breastmilk supplemented with formula. In contrast, the average distance was smallest between infants who were fed a mix of breastmilk and formula and those fed exclusively formula.
We were concerned that some of the subjects in the combination fed group may have been offered the breast in the first few days following delivery but were otherwise effectively exclusively formula fed, which may have driven the difference we observed between exclusively breast fed and combination fed infants in terms of between group differences. In fact, of subjects who were combination fed in our study, all but two were fed breastmilk for at least the first two weeks of life. To test the robustness of this finding in light of the possible effect of these two subjects, we repeated the UniFrac analysis after reassigning those two infants from the mixed feeding group to the exclusively formula fed group, and no qualitative differences in the results were observed (data not shown).
Individual taxon abundance by delivery mode and feeding method
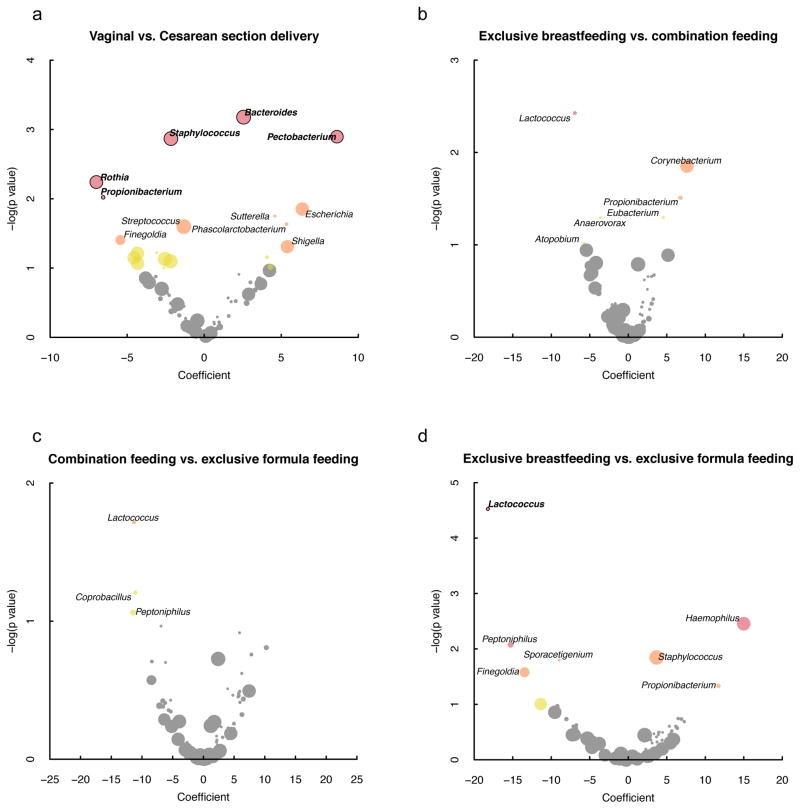
Vaginal delivery (vs. Cesarean section delivery) was associated with increased abundance of Bacteroides (p=0.0007, q=0.02) and Pectobacterium (p=0.001, q=0.02) and with decreased abundance of Staphylococcus (p=0.001, q=0.02), Rothia (p=0.006, q=0.07) and Propionibacterium (p=0.01, q=0.099), in infant stool, after adjustment for feeding method (figure 2a). Feeding was associated with differential abundance in Lactococcus (p<0.0001, q=0.002) which was depleted in exclusively breastfed infants compared with those who were exclusively formula fed (figure 2b). No taxa were significantly differentially abundant between subjects who were combination fed vs. exclusively formula fed or exclusively breastfed (figures 2c and 2d).
Fig. 2. Relationships between individual genus abundance and (a) delivery mode and (b–d) feeding method.
Positive coefficients indicate independent associations with vaginal delivery (a) or breastmilk exposure (b–d) after controlling for the other. Point colors correspond to adjusted p values of >0.1 (gray), ≤0.1 and >0.05 (yellow), ≤0.05 and >0.01 (orange), and ≤0.01 (red). Circles are sized according to relative log-ratio transformed abundance. Black borders/bold labels indicate genera that were significantly differentially abundant after controlling for the false discovery rate at a significance level of q=0.1. Note differences in axis scales.
Discussion
We characterized the intestinal microbiome of 102 six-week old infants and observed independent associations between stool microbial community composition, mode of delivery and feeding method. In healthy infants, the process of delivery is the initial encounter with microorganisms capable of colonizing the intestinal tract. In a previous study of 24 healthy women, vaginal microbiome composition became less diverse between the second and third trimesters of pregnancy, and just before delivery was enriched with Lactobacillus species, likely contributing to vertical transmission of these bacteria during vaginal birth 21. In a study of 10 newborns in Venezuela, within hours of delivery, the intestinal tracts of infants born vaginally were colonized by Lactobacillus and Prevotella, whereas infants delivered operatively acquired bacteria present on the mother’s skin and the hospital environment, such as Staphylococcus, Proprionibacterium and Corynebacterium 15. Our findings, based on a large group of six week old infants, indicate that Lactobacillus also contributes to the microbial environment of the gut, but to a lesser extent than Bifidobacteria, Bacteroides and Streptococcus.
Other studies have observed differences in older infants according to delivery mode. A study of 24 infants aged 3–4 months in Canada found that 2 out of 26 taxa evaluated were differentially abundant between vaginally and operatively delivered babies, including Bacteroides, which was depleted in Cesarean section delivered infants relative to those who were vaginally delivered 9. This result was also observed in a longitudinal study of 24 infants in Sweden, which reported that the depletion of Bacteroides in Cesarean section-delivered infants persisted until 12 months of age 13. Another longitudinal study of 75 infants in Singapore found that the acquisition of “normal” gut flora was delayed in infants born by Cesarean section 31.
Our study is the first to examine the contribution of delivery mode to infant intestinal microbiome composition in relationship to that of another important predictor of microbiome composition, infant diet. We found that delivery mode was more strongly associated with infant microbiome composition than was diet at six weeks. We observed differences in microbial community composition between vaginal and Cesarean section delivered infants that were comparable or slightly greater than the largest differences associated with feeding.
We observed an association between feeding method and microbiome composition that remained statistically significant even after adjusting for delivery mode. Though a few previous studies have found associations between infant feeding and intestinal microbiome composition 9–12,14, none has examined the relative contribution of combination feeding (breastmilk and formula) alongside exclusive formula or breastfeeding to overall microbial community composition. This is an important group to consider because combination feeding is common, for example, in the first few days in the hospital when lactogenesis II is delayed while mother’s breastmilk is becoming established, among mothers who have difficulty producing adequate milk and supplement their own milk with infant formula, or among mothers who are unable or choose not to pump breastmilk when separated from their babies. We found that the distinction between the microbial communities according to feeding method was largest between infants fed exclusively breastmilk and those fed either combination diets or exclusively formula. Infants fed both breastmilk and formula had intestinal microbial communities that were similar to those fed exclusively formula, and relatively distinct from those fed exclusively breastmilk. This finding offers new evidence to support the tenets of the World Health Organization’s Baby Friendly Hospital Initiative, which promotes exclusive breastmilk feeding beginning at birth in hospitals and birthing centers and the avoidance of formula supplementation unless deemed medically necessary (http://www.who.int/nutrition/topics/bfhi/en/). The findings in this study also provide new evidence for pediatricians as they provide guidance to breastfeeding mothers who may be considering incorporating formula into their infant’s diet, and may have implications for decisions around the use of donor human milk in cases when supplementation is needed.
There have been no long-term longitudinal studies of the effects of early feeding method on the microbiome, but early feeding has the potential for lasting effects on microbial community structure 32, and these impacts may be one mechanism for the health benefits of breastfeeding on childhood and lifelong health. Digestion and metabolism of nutrients are likely influenced by the intestinal microbiome 33, and there is a well-established connection between breastfeeding and lower risk of childhood and adult onset obesity likely mediated in part by the microbiome in early life (reviewed in 34). Oligosaccharides in breastmilk are thought to promote Bifidobacterium growth 35, and decreased Bifidobacterium in infancy has been related to an increased risk of being overweight at age 10 36. Many formulas are supplemented with prebiotics such as short chain galacto-oligosaccharides and long chain fructo-oligosaccharides that increase the overall representation of Bifidobacterium in the microbiome of formula fed infants, and similar to breastmilk, promote lactate and short chain fatty acid prevalence in the infant gut (reviewed in 37). Although we did not observe a significant association between increased abundance of Bifidobacterium and breastfeeding in our study, Bifidobacterium was present at greater abundance in exclusively breastfed infants compared with others; compared with combination fed subjects, this enrichment approached statistical significance before correction for multiple comparisons.
Our conclusions are limited by our study population, which was selected from a single cohort from the US and sampled at a single time point, and thus our findings may not be entirely generalizable to populations elsewhere or to different points in infant development. While ours is one of the largest studies examining the factors that shape the infant microbiome, our sample size of 102 subjects limited our statistical power which precluded stratified analyses for identifying any interactions between delivery mode and feeding method. In addition, while we were able to categorize feeding practices, the exact proportion of the diet that was made up of either breastmilk or formula and the exact timing of formula supplementation (e.g. in hospital after delivery versus beginning just prior to six weeks) was not considered. It is possible that infants who received formula supplementation only at birth were able to recover a microbiome that resembles that of an exclusively breast fed infant. A recent study highlighted infant nutrition as a major contributor to the early microbiota composition and function, with cessation of breastfeeding contributing the most fundamental shift in the composition of bacteria 8. A longitudinal study with more subjects would allow us to determine the temporal dynamics of the effects of feeding practices and changes therein, as well as the persistence of the effects of both feeding and delivery mode later in infancy. Additionally, exposures such as postnatal antibiotics were rare in this cohort and therefore subjects with antibiotic exposure were eliminated from analysis; in the future the evaluation of prenatal, peripartum, and postpartum antibiotic exposure and their role in the trajectory of microbiome development, and interrelationship with delivery mode and dietary exposures, will be important. Thus, our results will need to be replicated in larger, multi-center studies and in prospective analyses. While the UniFrac analyses we performed suggest independent associations between microbiome composition and both delivery mode and feeding method, the substantial overlap between the communities defined by both factors suggests that there are other important drivers of microbiome community composition that remain to be identified in future analyses.
In conclusion, understanding the patterns of microbial colonization of the intestinal tract of healthy infants is critical for determining the health impacts of specific, alterable early life risk factors and exposures. To this end, we have identified measurable differences in microbial communities in the intestinal tracts of infants according to their delivery mode and diet, with possible consequences for both short and long term health.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported in part by grants from the National Institutes of Health (NIEHS P01ES022832, NIEHS P20ES018175, NIGMS P20GM104416, and NLM K01LM011985) and the US Environmental Protection Agency (RD83459901 and RD83544201).
Footnotes
Author Contributions: Design and conduct of research performed by KLC, SFF, JCM and MRK; collection, management, analysis, and interpretation of data were conducted by AGH, JHM, HM, MS, JCM, KLC, and HL ; preparation, review, or approval of the manuscript were performed by AGH, MRK and JCM; all authors reviewed, revised, and participated in the decision to submit the manuscript for publication. JCM and AGH had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict of Interest Disclosures: None reported.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: Microbiology laboratory expertise and direction of Molly Housman, MS, of Geisel School of Medicine was invaluable. The authors are grateful to the children and families that made this study possible and to the staff of the New Hampshire Birth Cohort Study.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009 May;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng T, Elson CO. Adaptive immunity in the host-microbiota dialog. Mucosal Immunol. 2011 Jan;4(1):15–21. doi: 10.1038/mi.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010 Dec 24;330(6012):1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011 May 20;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011 Jun;23(3):353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thaiss CA, Zeevi D, Levy M, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014 Oct 23;159(3):514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 7.Madan JC, Farzan SF, Hibberd PL, Karagas MR. Normal neonatal microbiome variation in relation to environmental factors, infection and allergy. Curr Opin Pediatr. 2012 Dec;24(6):753–759. doi: 10.1097/MOP.0b013e32835a1ac8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Backhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host & Microbe. 2015 May 13;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013 Mar 19;185(5):385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz S, Friedberg I, Ivanov IV, et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012;13(4):r32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tannock GW, Lawley B, Munro K, et al. Comparison of the compositions of the stool microbiotas of infants fed goat milk formula, cow milk-based formula, or breast milk. Appl Environ Microbiol. 2013 May;79(9):3040–3048. doi: 10.1128/AEM.03910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012 Jun 14;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakobsson HE, Abrahamsson TR, Jenmalm MC, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014 Apr;63(4):559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 14.Fan W, Huo G, Li X, et al. Diversity of the intestinal microbiota in different patterns of feeding infants by Illumina high-throughput sequencing. World J Microbiol & Biotechnol. 2013 Dec;29(12):2365–2372. doi: 10.1007/s11274-013-1404-3. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blustein J, Attina T, Liu M, et al. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obesity. 2013 Jul;37(7):900–906. doi: 10.1038/ijo.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker E, Engelmann G, Findeisen A, et al. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Pediatr. 2010 Jun;125(6):e1433–1440. doi: 10.1542/peds.2009-2260. [DOI] [PubMed] [Google Scholar]
- 18.Cardwell CR, Stene LC, Joner G, et al. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia. 2008 May;51(5):726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 19.Kolokotroni O, Middleton N, Gavatha M, Lamnisos D, Priftis KN, Yiallouros PK. Asthma and atopy in children born by caesarean section: effect modification by family history of allergies - a population based cross-sectional study. BMC Pediatr. 2012;12:179. doi: 10.1186/1471-2431-12-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ip S, Chung M, Raman G, et al. Breastfeeding and maternal and infant health outcomes in developed countries. Evid Rep Technol Assess (Full Rep) 2007 Apr;(153):1–186. [PMC free article] [PubMed] [Google Scholar]
- 21.Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civardi E, Garofoli F, Mazzucchelli I, Angelini M, Manzoni P, Stronati M. Enteral nutrition and infections: the role of human milk. Early Hum Dev. 2014 Mar;90( Suppl 1):S57–59. doi: 10.1016/S0378-3782(14)70019-2. [DOI] [PubMed] [Google Scholar]
- 23.Mueller NT, Bakacs E, Combellick J, Grigoryan Z, Dominguez-Bello MG. The infant microbiome development: mom matters. Trends in Mol Med. 2015 Feb;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Nimwegen FA, Penders J, Stobberingh EE, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011 Nov;128(5):948–955. e941–943. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Farzan SF, Korrick S, Li Z, et al. In utero arsenic exposure and infant infection in a United States cohort: a prospective study. Environ Res. 2013 Oct;126:24–30. doi: 10.1016/j.envres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert-Diamond D, Cottingham KL, Gruber JF, et al. Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20656–20660. doi: 10.1073/pnas.1109127108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Degnan PH, Ochman H. Illumina-based analysis of microbial community diversity. ISME J. 2012 Jan;6(1):183–194. doi: 10.1038/ismej.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012 Aug;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Bittinger K, Charlson ES, et al. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012 Aug 15;28(16):2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dogra S, Sakwinska O, Soh SE, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio. 2015;6(1) doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fallani M, Amarri S, Uusijarvi A, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiol-Sgm. 2011 May;157(Pt 5):1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 33.Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011 Jul 1;94(1):58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson AL. Developmental origins of obesity: early feeding environments, infant growth, and the intestinal microbiome. Am J Hum Biol. 2012 May-Jun;24(3):350–360. doi: 10.1002/ajhb.22254. [DOI] [PubMed] [Google Scholar]
- 35.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci U S A. 2011 Mar 15;108( Suppl 1):4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luoto R, Kalliomaki M, Laitinen K, et al. Initial dietary and microbiological environments deviate in normal-weight compared to overweight children at 10 years of age. J Pediatr Gastroenterol and Nutr. 2011 Jan;52(1):90–95. doi: 10.1097/MPG.0b013e3181f3457f. [DOI] [PubMed] [Google Scholar]
- 37.Oozeer R, van Limpt K, Ludwig T, et al. Intestinal microbiology in early life: specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am J Clin Nutr. 2013 Aug;98(2):561S–571S. doi: 10.3945/ajcn.112.038893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.