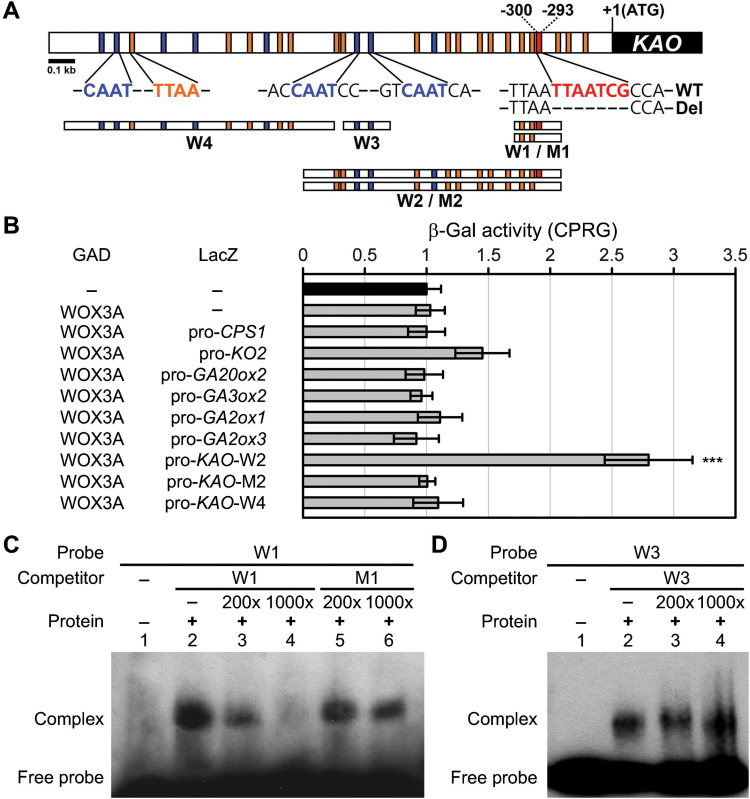
Fig. 6.
OsWOX3A protein directly binds to the promoter region of KAO.
(A) The locations of the W1, M1, W2, M2, W3, and W4 probes within the promoter region of KAO. The W1 probe includes the TTAATCG sequence, which is shown in red letters and by a red box. Reported core binding elements for WUS are indicated in blue and orange and by blue and orange boxes. (B) Analysis of OsWOX3A binding to the promoters of GA metabolic genes (CPS1, KO2, KAO, GA20ox2, GA3ox2, GA2ox1, and GA2ox3) using yeast one-hybrid assays. β-Galactosidase (β-Gal) activity was measured by liquid assay using chlorophenol red-β-D-galactopyranoside (CPRG). Each promoter binding activity is shown relative to the CPRG unit (104 ml−1 min−1) of the negative control that contains empty bait and prey plasmids (–), which is set as 1. Data are means ± SD from six independent colonies. Significant difference was determined by Student’s t-test (***P < 0.001). (C, D) EMSA showing His-OsWOX3A fusion protein binding to the KAO promoter. Oligonucleotides containing W1 (the KAO promoter binding site) or W3 (the KAO promoter with reported binding element for WUS), were used as the biotin-labelled probes. The W1 probe is shown in red letters in (A) and the M1 probe is shown with a dotted line in Del in (A). The negative control is indicated in Lane 1 in (C, D). Biotin-unlabelled W1, M1 (deleted version of W1), and W3 were used as the unlabelled competitors. The (+) presence or (−) absence of His-OsWOX3A fusion protein is indicated. These experiments were repeated more than three times with similar results.