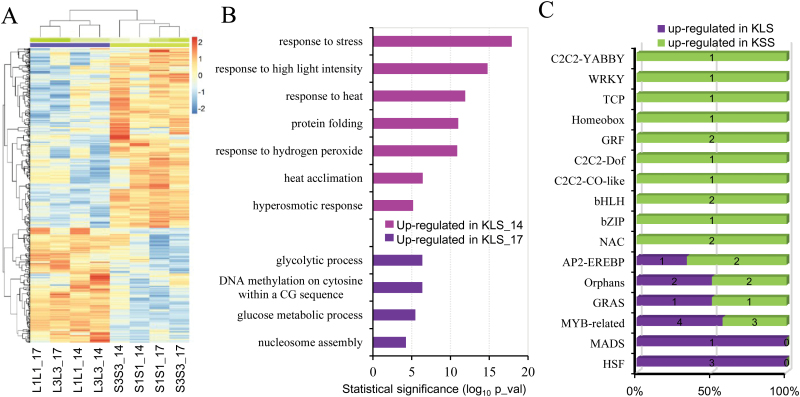
Fig. 4.
Differentially expressed genes (DEGs) between inbred lines derived from the KSS30 and KLS30 populations. (A) Heatmap of DEGs with 2-fold up- or down-regulation between KLS and KSS inbreds (KLS versus KSS) at 14 DAP and 17 DAP. (B) Significantly enriched Gene Ontology (GO) categories with the up-regulated genes in KLS inbred lines. (C) Distribution of transcription factor families among DEGs. The percentage on the x-axis was plotted as the number of differentially expressed transcription factors in each genotype out of all the differential transcription factors identified in the family. _14 and _17 indicate samples collected at 14 DAP and 17 DAP, respectively. S1S1, S3S3, L1L1, and L3L3 are inbred lines derived from KSS30 and KLS30, respectively.