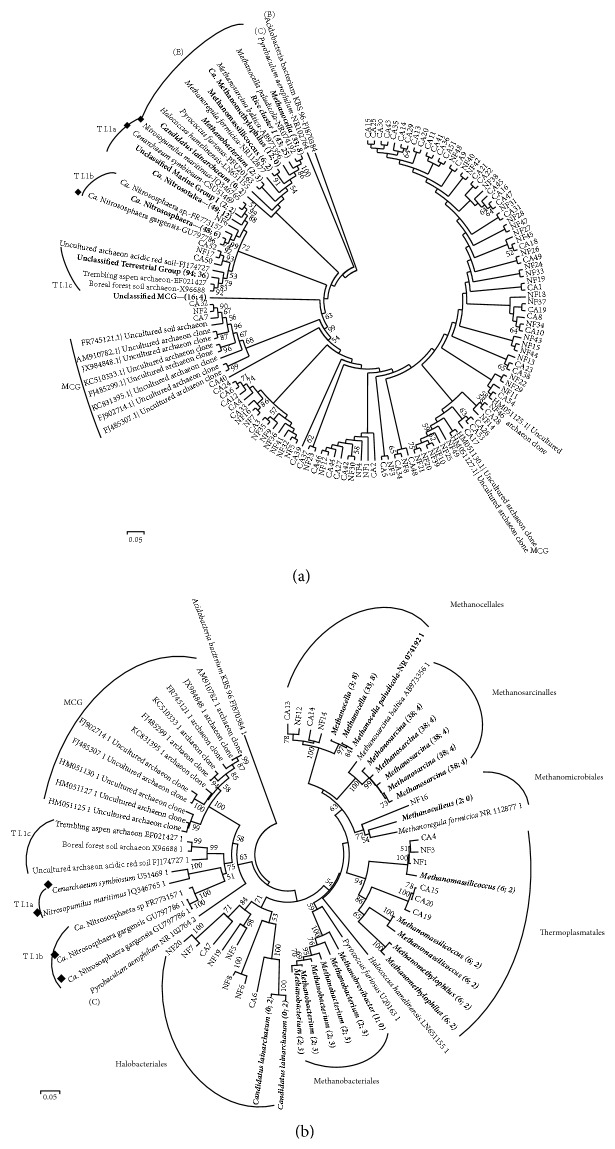
Figure 5.
Phylogenetic trees of (a) unclassified Thaumarchaeota 16S rRNA gene sequences from native forest soil and soil under oil palm cultivation and (b) 16S rRNA gene sequences classified as Euryarchaeota from native forest soil and soil under oil palm cultivation. The neighbor-joining method and Jukes-Cantor corrections were used with 1,000 bootstrap replicates. Bootstrap values above 50 are shown in the trees. A representative number of operational taxonomic units (OTUs) were selected at 3% dissimilarity by MOTHUR to avoid redundancies in the phylogenetic trees. Randomly chosen OTUs from native forest soil and from soil under oil palm cultivation were used in the construction of the trees, along with archaeal reference sequences (from GenBank) from the following groups: Acidobacteria (B), Crenarchaeota (C), Euryarchaeota (E), Miscellaneous Crenarchaeotic Group (MCG), Thaumarchaeota Group I.1a (T I.1a), Thaumarchaeota Group I.1b (T I.1b), and Thaumarchaeota Group I.1c (T I.1c). The AOAs are highlighted with the symbol ◆. Sampling site is indicated by “NF” (native forest) or “CA” (oil palm-cultivated area), followed by the number of the OTUs (NAxx or CAxx). OTU sequences classified to at least the class level are shown in bold, followed by two numbers in parentheses indicating the number of sequences of that OTU detected in native forest soil and the number detected in the oil palm-cultivated area. The scale bar represents the 5% estimated sequence divergence.