Abstract
Myrosinases are β-thioglucoside glucohydrolases and serve as defense mechanisms against insect pests and pathogens by producing toxic compounds. AtTGG6 in Arabidopsis thaliana was previously reported to be a myrosinase pseudogene but specifically expressed in pollen. However, we found that AlTGG6, an ortholog to AtTGG6 in A. lyrata (an outcrossing relative of A. thaliana) was functional, suggesting that functional AtTGG6 alleles may still exist in A. thaliana. AtTGG6 alleles in 29 A. thaliana ecotypes were cloned and sequenced. Results indicate that ten alleles were functional and encoded Myr II type myrosinase of 512 amino acids, and myrosinase activity was confirmed by overexpressing AtTGG6 in Pichia pastoris. However, the 19 other ecotypes had disabled alleles with highly polymorphic frame-shift mutations and diversified sequences. Thirteen frame-shift mutation types were identified, which occurred independently many times in the evolutionary history within a few thousand years. The functional allele was expressed specifically in pollen similar to the disabled alleles but at a higher expression level, suggesting its role in defense of pollen against insect pests such as pollen beetles. However, the defense function may have become less critical after A. thaliana evolved to self-fertilization, and thus resulted in loss of function in most ecotypes.
Keywords: myrosinase, β-thioglucosidase, AtTGG6, AlTGG6, evolution, pollen specific, frame-shift mutation
1. Introduction
Plant myrosinase (β-thioglucoside glucohydrolase, EC 3.2.1.147) belongs to one of the currently 135 families in the glycoside hydrolase superfamily [1,2]. It is a unique class of glycosidases by catalyzing the hydrolysis of S-linked glucosides, glucosinolates (anionic 1-thio-β-d-glucoside N-hydroxysulphates). Glucosinolates occur mainly in Brassicale plants [3], and release toxic compounds when hydrolyzed by myrosinase. Thus, myrosinase plays important roles in the defense system of cruciferous plants [4,5].
All crucifers analyzed so far have multiple forms of myrosinases. In oilseed rape (Brassica napus) and other Brassica species, approximately 25–30 myrosinase isoforms may be present [6,7,8]. These myrosinases were divided into three subgroups (MA, MB, and MC) based on their biochemical properties and sequence similarities [8]. Arabidopsis thaliana was shown to have six myrosinase genes AtTGG1–6, among which AtTGG1 and AtTGG2 were functional and expressed in above ground tissues [4,9], while AtTGG4 and AtTGG5 were expressed in roots [10,11]. Interestingly, AtTGG3 and AtTGG6 were disabled in tested ecotypes, but specifically expressed in anthers [12,13]. Carica papaya had at least three myrosinase genes [14,15], which were clustered together with AtTGG4 and AtTGG5 in a new subgroup of myrosinases Myr II, while MA, MB, and MC myrosinases together with AtTGG1–3 were clustered in another subgroup Myr I by phylogenetic analysis [14,15]. Myr I and II were also different by their exon/intron organizations, in which Myr I had 12 exons and used an unusual intron splicing border GC/AG for intron 1 [16], while Myr II had 13 exons and used the unusual intron splicing border GC/AG for intron 10 and the normal border GT/AG for intron 1. However, papaya myrosinase genes did not use any unusual splicing border [14,15].
The anther specific expression of the disabled AtTGG6 in A. thaliana suggested its possible function in the evolutionary history [12], and moreover, database searches in Arabidopsis lyrata genome identified a functional ortholog AlTGG6, which suggested that functional alleles of AtTGG6 may still exist in A. thaliana ecotypes. In this study, we report identification of surprisingly high frequency of functional alleles and highly polymorphic frame-shift mutations in the non-functional AtTGG6 alleles.
2. Results
2.1. Identification of Functional Alleles of the AtTGG6 Locus in Arabidipsis Thaliana
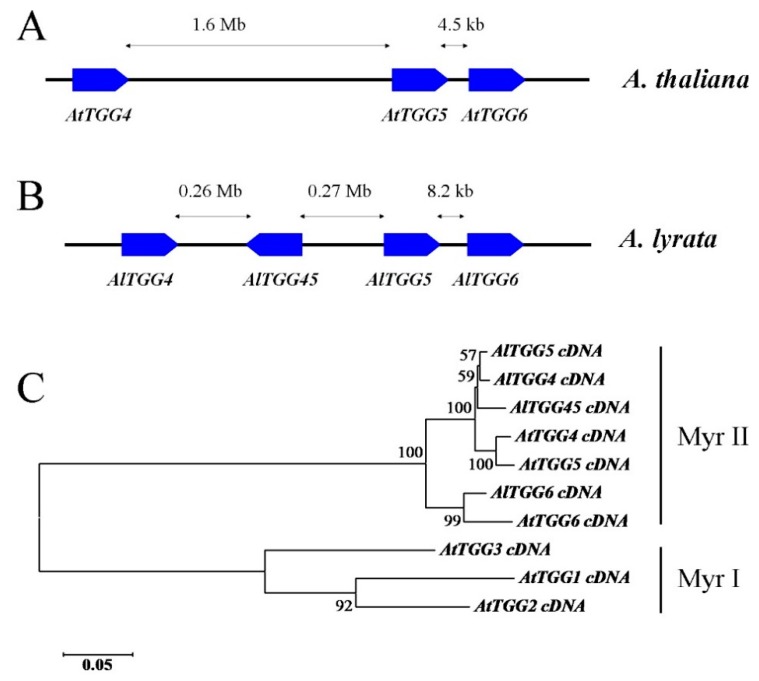
Myrosinase gene AtTGG6 is a Myr II member gene in Arabidopsis thaliana and was previously reported to be disabled in five tested ecotypes [12]. Interestingly, it was predominantly expressed in pollen [12]. Blast searches in Arabidopsis lyrata subsp. lyrata genome resulted in four Myr II type myrosinase gene members designated here as AlTGG4, AlTGG5, AlTGG6, and AlTGG45. They locate in the chromosome with an organization similar to the Myr II genes AtTGG4, AtTGG5, and AtTGG6 in A. thaliana (Figure 1A), except that AlTGG4 and AlTGG5 are separated by the inverted gene AlTGG45 (Figure 1B). All the four A. lyrata myrosinase genes have the same exon/intron organizations as the Myr II members with 13 exons and the unusual intron splicing border GC/AG for intron 10, suggesting that this intron splicing border had existed before the two species were diverged. AlTGG4, AlTGG5, and AlTGG6 encode preproteins of 510, 510, and 512 residues, respectively, and are likely functional, while AlTGG45 is likely non-functional due to a 2412 bp insertion in exon 3 (GenBank accession number KU301859).
Figure 1.
Chromosomal locations and phylogenetic analysis of Myr II members in Arabidopsis lyrata and A. thaliana. (A) Chromosomal locations of Myr II genes in A. thaliana; (B) Chromosomal locations of Myr II genes in A. lyrata; (C) Phylogeny inferred from cDNAs of Myr II members in A. thaliana and A. lyrata, using Myr I myrosinases AtTGG1 (At5g26000), AtTGG2 (At5g25980), and AtTGG3 (At5g48375) as outgroups. Note: AtTGG4 (At1g47600) and AtTGG5 (At1g51470) cDNA sequences were from ecotype Col-0, AtTGG6 (At1g51490) sequence was from ecotype Tsu-1 (GenBank accession number KU301834). AlTGG cDNA sequences were obtained by annotating a genomic scaffold (NW_003302555) in A. lyrata, and partially confirmed by sequencing the PCR-amplified cDNAs. The sequences were deposited in the GenBank database under accession numbers KU301856, KU301857, KU301858, and KU301859 for AlTGG4, AlTGG5, AlTGG6, and AlTGG45, respectively.
Phylogenetic analysis indicated that AlTGG4, AlTGG5, and AlTGG45 were clustered together with AtTGG4 and AtTGG5 in the Myr II clade, but they were subdivided by species (Figure 1C), suggesting that TGG4 and TGG5 in the genus Arabidopsis were duplicated after the two species had diverged. However, AtTGG6 and AlTGG6 were clustered in another subgroup in the Myr II clade (Figure 1C), which suggested that AlTGG6 is an ortholog to AtTGG6, and functional AtTGG6 alleles may still exist in some ecotypes of A. thaliana.
AtTGG6 genomic DNAs were amplified and sequenced in 29 A. thaliana ecotypes. In contrast to our expectation, as many as 10 ecotypes contained functional alleles. cDNAs of the functional alleles in two representative ecotypes Tsu-1 and Ty-0 were cloned and sequenced. They encoded preproteins of 512 residues in which seven residues were different between them, and they shared approximately 95% amino acid identities with AlTGG6. A secretion signal of 22 residues was identified by the SignalP 4.0 server [17], and the predicted molecular mass after signal excision was 55.7 kD, and pI was 9.4.
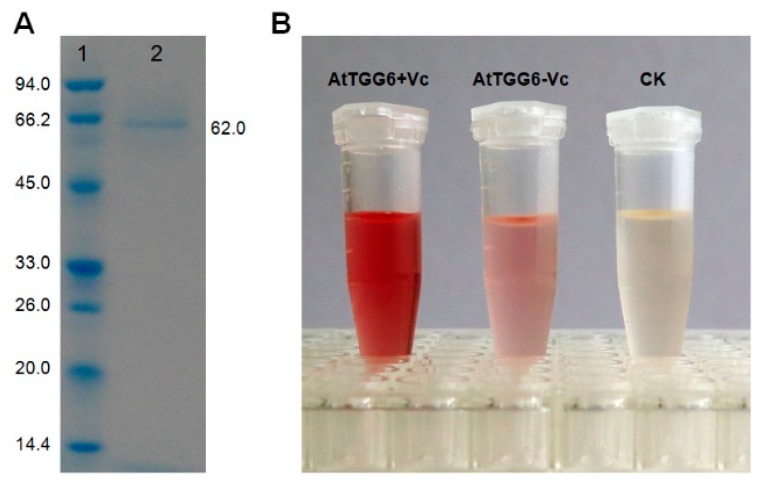
The AtTGG6 from ecotype Tsu-1 was overexpressed in Pichia pastoris. The purified recombinant protein had a molecular weight of approximately 62 kD (Figure 2), higher than the predicted mass, possibly due to glycosylation, which is common in myrosinase [14,15,18]. Myrosinase activity of the purified protein was confirmed by its ability to release glucose from sinigrin, a common myrosinase substrate (Figure 2). The production of glucose was visualized using a glucose test reagent containing glucose oxidase and 4-aminoantipyrin (GOD-PAP). Myrosinase activity of the recombinant AtTGG6 was activated by adding 0.8 mM ascorbic acid in the reaction mixture (Figure 2). Ascorbate (Vc) activation is a well-known plant myrosinase characteristic [15,19,20,21].
Figure 2.
Myrosinase activity of recombinant AtTGG6 from Arabidopsis ecotype Tsu-1. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of recombinant protein overexpressed in Pichia pastoris GS115; Lane 1, molecular weight marker, the mass of each band in kilodalton (kD) is shown to the left; Lane 2, purified recombinant AtTGG6; (B) Myrosinase activity of purified AtTGG6 as visualized using a glucose test reagent; AtTGG6 + Vc, AtTGG6 (50 ng) plus 0.8 mM ascorbic acid (Vc); AtTGG6 − Vc, AtTGG6 (50 ng) only, Vc not added; CK, AtTGG6 (50 ng) disabled by heating at 95 °C for 5 min before use.
The exon/intron structure of the functional AtTGG6 was the same as the Myr II members with 13 exons and the unusual GC/AG splicing border for intron 10. When the sequences of the functional alleles were aligned with the disabled allele in A. thaliana ecotype Col-0, all the frame-shift mutations in Col-0 previously predicted were confirmed [12] (Figure S1).
2.2. Highly Active Frame-Shift Mutations in AtTGG6 Alleles
The AtTGG6 locus is a hot spot for frame-shift mutations. Among the 19 disabled alleles, a total of 13 frame-shift mutation types were identified, which were designated as Mu1 to Mu13 (Table 1). The mutations include insertion and deletion mutations (InDels) in exons, such as one base insertion in exon 3 (Mu3), exon 6 (Mu4), and exon 11 (Mu11); two base insertion in exon 1 (Mu1) and exon 7 (Mu5); one base deletion in exon 12 (Mu13); four base deletion in exon 9 (Mu6); 14 base deletion in exon 9 (Mu7); 17 base deletion in exon 11 (Mu8) and exon 12 (Mu12); and 208 base deletion from intron 2 to exon 4 (Mu2). Mutations in the exon/intron splicing border, such as 3′-splicing border mutation from AG to AT (Mu9) and/or to GG (Mu10) were also identified, which resulted in splicing failure for intron 10. The failure in splicing was confirmed by sequencing the cDNAs in ecotype Gre-0 that contained Mu9 and Stw-0 that contained Mu10 (cDNA sequences have been deposited in GenBank at accession numbers KU301844 and KU301851, respectively). The ecotypes and relevant frame-shift mutations are listed in Table 2.
Table 1.
Frame-shift mutations identified in AtTGG6 alleles in Arabidopsis thaliana ecotypes.
| Mutation | Location |
|---|---|
| Mu1 | Two base insertion at +155 * in exon 1 |
| Mu2 | 208 base deletion from +437 in intron 2 to +644 in exon 4 |
| Mu3 | one-base insertion at +481 in exon 3 |
| Mu4 | one-base insertion at +1007 in exon 6 |
| Mu5 | Two-base insertion at +1158 in exon 7 |
| Mu6 | four-base deletion from +1739 to +1742 in exon 9 |
| Mu7 | 14 base deletion from +1804 to +1817 in exon 9 |
| Mu8 | 17 base deletion from +2111 in intron 10 to +2127 in exon 11 |
| Mu9 | 3’ splicing border mutation from AG to AT for intron 10 |
| Mu10 | 3’ splicing border mutation from AG to GG for intron 10 |
| Mu11 | One-base insertion at +2121 in exon 11 close to 3’ splicing border of intron 10 |
| Mu12 | 17 base deletion from +2311 to +2327 in exon 12 |
| Mu13 | One nucleotide deletion at +2348 in exon 12, 20 nucleotides after Mu12 |
* The number was counted from the first nucleotide of the start codon in AtTGG6 gene of ecotype Tsu-1 (GenBank accession number KU301834).
Table 2.
Distribution of frame-shift mutations in AtTGG6 alleles among Arabidopsis ecotypes.
| Ecotype | Mu1 | Mu2 | Mu3 | Mu4 | Mu5 | Mu6 | Mu7 | Mu8 | Mu9 | Mu10 | Mu11 | Mu12 | Mu13 | Functionality |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aa-0 | + | − | − | − | − | − | − | − | − | − | − | − | − | disabled |
| Ag-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Ba-1 | − | − | − | − | − | − | − | − | − | − | − | − | + | disabled |
| Col-0 | + | − | − | + | − | + | − | + * | − | − | − | + | − | disabled |
| Col-4 | + | − | − | + | − | + | − | + * | − | − | − | + | − | disabled |
| Col-J | + | − | − | + | − | + | − | + | − | − | − | + | − | disabled |
| Cvi-0 | + | − | − | + | − | + | − | + | − | − | − | + | − | disabled |
| Bsch-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Edi-0 | − | − | − | − | − | − | − | − | − | − | − | − | + | disabled |
| Gel-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Gre-0 | − | − | + | − | − | − | − | − | + * | − | − | − | − | disabled |
| JM-1 | − | − | − | + | − | − | − | − | − | − | − | − | − | disabled |
| JM-2 | − | − | − | + | − | − | − | − | − | − | − | − | − | disabled |
| Kas-1 | + | − | − | − | − | − | − | − | − | − | − | − | − | disabled |
| La-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Ler-1 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Mir-0 | − | − | − | − | + | − | − | − | − | − | + * | − | − | disabled |
| Mz-0 | − | − | − | − | − | − | − | − | − | + | − | − | − | disabled |
| Mr-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Oy-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Pr-0 | + | + | N/A ** | − | − | − | + | − | − | − | − | − | − | disabled |
| Stw-0 | + | − | − | − | − | − | − | − | − | + * | − | − | − | disabled |
| Ts-6 | − | − | − | − | + | − | − | − | − | − | − | − | − | disabled |
| Tsu-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Tul-0 | − | − | + | − | − | − | − | − | + | − | − | − | − | disabled |
| Ty-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Van | − | − | − | − | − | − | − | − | − | + | − | − | − | disabled |
| Ws-0 | − | − | + | − | − | − | − | − | − | − | − | − | − | disabled |
| Wil-0 | − | − | − | − | − | − | − | − | − | − | − | − | − | functional |
| Frequency | 8 | 1 | 3 | 6 | 2 | 4 | 1 | 4 | 2 | 3 | 1 | 4 | 2 | - |
* Confirmed by sequencing cDNA sequences amplified with RT-PCR method; ** Not applicable, since Mu2 in ecotype Pr-0 is a large deletion that spans the Mu3 region.
The predicted nine base deletion in the intron 10/exon 11 junction in ecotype Col-0 in our previous report [12] was actually 17 bases (Mu8) according to current alignment with the functional alleles (Figure S1), which includes four bases in intron 10 and 13 bases in exon 11. Four ecotypes (Col-0, Col-4, Col-J, and Cvi-0) contained this mutation, which resulted in retaining of intron 10 in the mRNA in ecotype Col-0 and Col-4 as confirmed by sequencing their cDNAs (GenBank accession numbers KU301839 and KU301840).
Mu1 had the highest frequency, and eight out of the 19 disabled alleles contained this mutation, while Mu2, Mu7, and Mu11 were identified in only one allele (Table 2). The 3′ splicing border region of the unusual intron 10 is one of the hottest targets for disable mutation. Four types of mutations (Mu8, Mu9, Mu10, and Mu11) were found in this region with a total of 10 events in 10 distinct ecotypes. All these events resulted in failure of intron splicing as revealed by sequencing the cDNAs of representative ecotypes (Table 2). It is not known whether there is a relationship between the rare intron splicing border and the mutation rate.
2.3. Genetic Diversity and Evolutionary History of AtTGG6
AtTGG6 was highly diverged according to the genetic diversity parameters calculated with DnaSP5 software [22]. The overall sequence identities of AtTGG6 alleles were only 85.4%. The haplotype diversity was 0.973 with 109.5 InDel sites and 36.5 single nucleotide polymorphic (SNP) sites per kb DNA (Table 3).
Table 3.
Genetic diversity in AtTGG6 alleles in 29 Arabidopsis thaliana ecotypes.
| Diversity Parameters | Genomic Gene | Exons | Introns | Functional Alleles | Nonfunctional Alleles |
|---|---|---|---|---|---|
| Sequence identities | 85.4% | 86.7% | 80.5% | 97.5% | 86.0% |
| Haplotype diversity | 0.973 ± 0.018 | 0.968 ± 0.019 | 0.948 ± 0.024 | 0.911 ± 0.077 | 0.959 ± 0.036 |
| Nucleotide diversity per site | 0.00758 ± 0.00085 | 0.00593 ± 0.00055 | 0.01051 ± 0.00167 | 0.00770 ± 0.00082 | 0.00666 ± 0.00140 |
| θw per site | 0.01043 ± 0.00336 | 0.00968 ± 0.00323 | 0.01199 ± 0.00405 | 0.00691 ± 0.00291 | 0.01027 ± 0.00364 |
| SNP sites per kb | 36.485 | 34.282 | 39.792 | 5.605 | 32.030 |
| InDel sites per kb | 109.456 | 98.318 | 154.844 | 19.432 | 107.635 |
| Total aligned length | 2686 | 1546 | 1156 | 2676 | 2685 |
The diversity of exons and introns was calculated separately. Their haplotype diversity was similar, but the nucleotide diversity of introns was significantly higher than that of exons, and there were more InDels and SNPs in the introns (Table 3). These results suggested that there had been a selection pressure for a functional AtTGG6 in the evolutionary history. This hypothesis was also supported by the diversity parameters of the functional and the disabled alleles calculated separately. The haplotype diversity, the population mutation rate θw, InDel sites, and SNP sites were all higher in the disabled alleles than those in the functional alleles, except that the nucleotide diversity per site in the functional alleles was higher than that in the disabled alleles (Table 3). The higher nucleotide diversity in the functional alleles may have resulted from the accumulation of synonymous mutations in the evolutionary history. The disabled alleles were evolved from the functional alleles, and thus had a shorter evolutionary history compared to the functional alleles, and had accumulated less synonymous mutations. However, frame-shift mutations were accumulated in the population of the disabled alleles due to the lack of selection pressure after mutation.
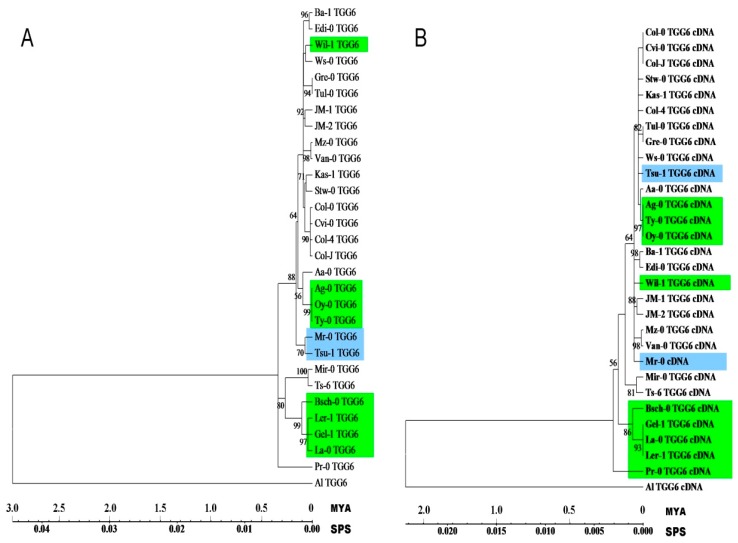
Phylogenetic analysis of AtTGG6 genomic and cDNA sequences of the 29 alleles revealed that the disabled alleles dispersed in different clades represented by different functional alleles (Figure 3), suggesting that disable mutation events happened many times independently in the evolutionary history, and most of the events happened within 50 thousand years when genomic DNA was used to estimate the timing (Figure 3A), and the timing was narrowed down to a few thousand years when cDNA was used (Figure 3B), and this mutation rate may have been accelerated in human greenhouses where A. thaliana is grown for many generations a year.
Figure 3.
Phylogenetic analysis of AtTGG6 genomic (A) and cDNA (B) sequences by Maximum Likelihood method. The evolutionary history was inferred based on the Tamura–Nei model [23]. The bootstrap values over 50% are shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically. The tree is drawn to scale, with branch lengths measured in million years (MYA) and substitutions per site (SPS). Evolutionary analyses were conducted in MEGA6 [24]. Functional alleles are highlighted in green or blue. The two functional alleles in Tsu-1 and Mr-0 which are clustered different in genomic and cDNA phylogenies are highlighted in blue.
2.4. Expression Pattern of the Functional Allele in Arabidopsis thaliana
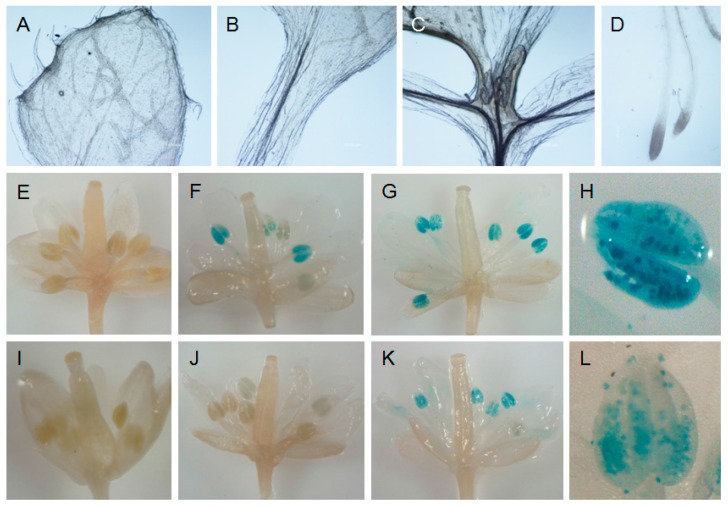
The promoters of the functional (from Tsu-1) and the disabled (from Col-0) AtTGG6 were respectively fused with GUS gene and transformed into ecotype Col-0. GUS assay revealed that the functional AtTGG6 was expressed predominantly in pollen, but not expressed in other tissues of the flower, neither expressed in leaves, flower stalk, and roots (Figure 4A–H). This expression pattern was similar to the disabled allele in Col-0 (Figure 4I–L). However, the expression level of the functional allele was higher than that of the disabled allele as visualized by darkness of blue color. These results indicated that the functional AtTGG6 was possibly responsible for protection of pollen against pest insects such as pollen beetles, which caused severe crop loss in oilseed rape (Brassica napus L.) in some regions of the world [25].
Figure 4.
Expression pattern of functional (A–H) and disabled (I–L) alleles of AtTGG6 in Arabidopsis thaliana. Functional Prom::GUS and disabled Prom::GUS were transformed into Col-0, and stained with X-gluxuronide solution. (A) Rosette; (B) stem; (C) leaves; and (D) root of transgenic plant with functional Prom::GUS; (E) a flower bud opened with a needle showing no GUS staining; (F) a flower one day before flowering opened with a needle showing the early expression; (G) a fully opened flower showing the high expression level; (H) an anther showing predominant expression in pollen; (I) a flower bud of disabled Prom::GUS opened with a needle showing no GUS staining; (J) a flower of disabled Prom::GUS one day before flowering showing weak expression; (K) a fully opened flower of disabled Prom::GUS showing expression at moderate level; (L) an anther showing disabled AtTGG6 predominantly transcribed in pollen.
3. Discussion
AtTGG6 was previously suggested to be a pseudogene after sequencing the genes in five ecotypes [12]. Surprisingly, ten out of 29 ecotypes were found to have functional alleles in this study, and the expression pattern of the functional allele was similar to that of the disabled allele, which suggested that the functional AtTGG6 was responsible for resistance against pollen herbivores in the evolutionary history based on known functions of myrosinases [8,26,27]. However, the necessity to defend pollen in A. thaliana may have become weaker than its outcrossing relative A. lyrata, after A. thaliana had become an autogamous species and can pollinate before the flower is open by disabling its self-incompatibility (SI) genes. SI genes still exist widely in crucifers [28,29,30]. Therefore, the strength or efficacy of natural selection for a functional AtTGG6 decreased, and thus, disabled alleles survived, which may be one of the explanations for the high frequency of frame-shift mutations in AtTGG6. Gene expression patterns have been hypothesized not only to be the major determinants of nonsynonymous variation rate among genes, but also a critical determinant of gene retention [31,32]. A second factor that may have influenced the mutation of AtTGG6 may be related to self-fertilization itself, which resulted in a large reduction in the effective rate of recombination and a corresponding decline in effective population size, and accelerated mutation [32,33]. A third factor that may have accelerated the mutation of AtTGG6 may be the singleton nature of this gene in the genome. AtTGG6 was localized closely to AtTGG4 and AtTGG5 in the chromosome (Figure 1A), however, phylogenetic analysis indicated that AtTGG4 and AtTGG5 were subdivided with AlTGG4, AlTGG5, and AlTGG45 in A. lyrata by species in the Myr II clade (Figure 1C), suggesting that TGG4 and TGG5 in the genus Arabidopsis were duplicated recently, while TGG6 was a singleton, more diverged, and was evolved before A. thalina and A. lyrata were separated into two species (Figure 1C). Singletons had a higher frequency of evolutionary rate than duplicated genes [31].
Frame-shift mutation was a major force to drive AtTGG6 to lose function. We observed highly active frame-shift mutations in the AtTGG6 locus. A total of 13 frame-shift mutation types were identified in 29 ecotypes (Table 2). These disabled alleles were dispersed in different clades in the phylogenetic tree (Figure 3), suggesting that AtTGG6 was disabled many times independently in the evolutionary history. It is well known that myrosinase use GC/AG alternative intron splicing border [12,16]; Myr I and Myr II member genes use GC/AG for intron 1 and intron 10, respectively. Here, all the AtTGG6 alleles were found to use GC/AG for intron 10, except for the specific splicing mutants. Interestingly, the unusual intron splicing border region in AtTGG6 gene was a hot spot for frame-shift mutations (Table 2). A total of 10 frame-shift mutation events were identified in this region, including the mutations of splicing border from GC/AG to GC/AT and GC/GG. All these events resulted in failure of intron splicing as revealed by sequencing the cDNA of representative ecotypes. It is not known whether there is a relationship between the rare intron splicing border and the mutation frequency. Alternative splicing is common in plants [34,35]. However, alternative transcripts of AtTGG6 were not obtained and the GC/AT and GC/GG splicing borders in AtTGG6 were also proven not functional by RT-PCR analysis.
The functional alleles of AtTGG6 are retained in some ecotypes possibly due to insufficient evolutionary time, since self-fertilization in A. thaliana occurred just a million years ago [30], after A. thaliana was diverged from its outcrossing relative A. lyrata approximately 5 million years ago [28,32], and from Brassica species 15 to 20 million years ago [36]. Through analysis of sequence diversity and phylogenetic analysis, frame-shift mutations of AtTGG6 may have happened many times in the evolutionary history, and most mutations happened only a few thousand years ago (Figure 3), some may have even occurred recently in greenhouses.
4. Experimental Section
4.1. Plant Materials and Growth Conditions
Most ecotypes of A. thaliana are obtained from the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University (Columbus, OH, USA). Some ecotypes are old stocks in our lab. Their accession numbers and origins are listed in Table 4. The plants were grown in a culture room under 16/8 h photoperiod, 200 μmol·m−2·s−1 light intensity, and 20 °C.
Table 4.
Ecotypes used in this research.
| Ecotypes | ABRC Stock Number | Location |
|---|---|---|
| Aa-0 | CS937 | Aua/Rhon, Germany |
| Ag-0 | CS936 | Argentat ,France |
| Ba-1 | CS952 | Blackmount, United Kingdom |
| Bsch-0 | CS1002 | Buchschlag/Frankfurt am Main, Germany |
| Col-0 | CS6673 | Columbia, SC, USA |
| Col-4 | CS933 | Columbia, SC, USA |
| Cvi-0 | CS1096 | Cape Verde Islands, Cape Verde |
| Edi-0 | CS1122 | Edinburgh, United Kingdom |
| Gel-1 | CS28279 | Geleen, Netherland |
| Gre-0 | CS1210 | Greenville, MI, USA |
| Kas-1 | CS903 | Kashmir, India |
| La-0 | CS1298 | Landsberg/Warthe, Germany |
| Ler-1 | CS1642 | Landsberg, Germany |
| Mir-0 | CS1378 | Miramare/Trieste, Italy |
| Mr-0 | CS1372 | Monte/Tosso, Italy |
| Mz-0 | CS1382 | Merzhausen/Ts, Germany |
| Oy-0 | CS1436 | Oystese, Norway |
| Pr-0 | CS1474 | Frankfurt-Praunheim, Germany |
| Stw-0 | CS1538 | Stobowa/Orel, Russia |
| Tu1-0 | CS1570 | Turk Lake, MI, USA |
| Ts-6 | CS1561 | Tossa del Mar, Spain |
| Tsu-1 | CS1640 | Tsushima, Japan |
| Ty-0 | CS1572 | Taynuilt, United Kindom |
| Van-0 | CS1584 | Vancuuver, Canada |
| Wil-1 | CS1594 | Wilna/Litauen, Russia |
| Ws-0 | CS1602 | Wassilewskija, Russia |
| Col-J | - | Derived from Col-0 |
| JM-1 | - | Lab stock, unknown origin |
| JM-2 | - | Lab stock, unkown origin |
ABRC: Arabidopsis Biological Resource Center.
4.2. DNA Isolation and Cloning of AtTGG6 Genomic Genes
Leaves collected from the ecotypes were grounded in liquid nitrogen. DNA was isolated with a Universal Genomic DNA Extraction Kit Ver.3.0 (TaKaRa Biotechnologies, Dalian, China). Full length genomic gene of AtTGG6 was amplified with gene specific primers G6F1 (5′ ACC CGC TGA AAA GCT CCA TCA A 3′) and G6R1 (5′ GGC TTC CAC TTA TTT TGC AAT GAA CC 3′) and sequenced at Shanghai ShineGene Molecular Biotechnology Co., Ltd., Shanghai, China.
4.3. Isolation of mRNA, RT-PCR and Sequence Analysis
Plant samples of representative ecotypes were grounded in liquid nitrogen. Total RNA was isolated with 3S Trizol Total RNA isolation reagents (Shenergy Biocolor Bioscience & Technology Company, Shanghai, China). mRNA was then purified from the total RNA with an Oligotex Direct mRNA purification kit (Qiagen GmbH, Hilden, Germany). Moloney Murine Leukemia Virus (M-MuLV) reverse transcriptase (TaKaRa Biotechnologies, Dalian, China) was used to generate first strand cDNA. cDNA fragments of AtTGG6 were amplified with gene specific primers G6F1 and G6R1 and cloned in pMD19-T vector (TaKaRa Biotechnologies, Dalian, China). Both the PCR fragments and the clones were sequenced at ShineGene, Shanghai. The sequences were analyzed with MacVector software (Version 13.5.5, Oxford Molecular, Oxford, UK).
4.4. Overexpression of AtTGG6 in Pichia Pastoris
A full length cDNA clone containing the correct AtTGG6 sequence of ecotype Tsu-1 was selected to construct an intracellular expression vector. The coding region was re-amplified with primers HNP38 (5′ GGG TGA TCA GCA TGG CAA TTC CAA AAG CTC ACT ACT CT 3′) and HNP39 (5′ GGG CCT AGG CTA GTG GTG ATG GTG ATG GTG TTT TGC AAT GAA CCT AGA GAA CCA TTT TCC A 3′), using proofreading DNA polymerase LA Taq (TaKaRa Biotechnologies). The PCR reaction was performed for 30 cycles, at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 3 min and 30 s. The product was digested with Bcl I and Avr II, and ligated into an intracellular expression vector pPIC3.5 (Invitrogen, San Diego, CA, USA) that was previously digested with Bam HI and Avr II. The ligated product was transformed into Escherichia coli by electroporation. The recombinant plasmid with correct sequence was chosen to transform P. pastoris strain GS115 (Invitrogen) as previously described [14].
4.5. Purification of Recombinant AtTGG6 and Analysis of Myrosinase Activity
A single P. pastoris colony with high myrosinase activity was used to initiate large scale cultures as described previously [14]. The production of recombinant myrosinase was induced by addition of 0.5% methanol each day for five days. The his-tagged recombinant protein was isolated and purified with an 1-mL BioRad profinity cartridge as described previously [14]. The purity and molecular weight of recombinant enzyme was examined with a 4%–10% Bis–Tris SDS-PAGE gel and stained with Coomassie brilliant blue. Myrosinase activity was tested by its ability to release glucose from commonly used substrate sinigrin (Sigma Chemical Co., St. Louis, MO, USA). Glucose produced in the reaction was visualized by reaction with a glucose test reagent (Shanghai Rongsheng Biotech Co., Ltd., Shanghai, China) that is widely used to measure blood glucose level in hospitals.
4.6. Construction of AtTGG6 Prom::GUS Expression Vector, Transformation, and GUS Staining
The promoter of AtTGG6 was amplified as previously described with primers HNP56 (5′ GGA CTT TGG ATC ACC ATT AAG CTT GCA G 3′) and HNP57 (5′ GTT GGA TCC GCT TTC TTT TTG CGT TTC TTG ATG GAG 3′) [12] from ecotype Tsu-1 and Col-0. The product was digested with Hind III and Bam HI, and inserted into the pBI121 vector pre-digested with Hind III and Bam HI. The recombinant plasmids containing AtTGG6 Prom::GUS were transformed into E. coli and screened by restriction digestion and sequencing. The correct expression vectors were transformed into Agrobacterium tumefaciens C58 by electroporation, and were further transformed into Arabidopsis ecotype Col-0 using vacuum infiltration [37]. Transformants were screened on MS medium containing 40 mg·L−1 kanamycin. The expression of GUS gene was detected as described [38].
4.7. Analysis of DNA Sequences and Genetic Diversity
Sequences were assembled using the Assembler program incorporated in the MacVector 13.5.5 software (MacVector, Inc.). The sequences of AtTGG6 alleles were deposited in GenBank under accession numbers (KU301827–KU301855; Table S1). The Myr II member genes in A. lyrata were submitted in GenBank with accession numbers KU301856–KU301859 (Table S1). The exon/intron organizations and the frame-shift mutations in the AtTGG6 alleles in different ecotypes were identified by aligning the genomic DNA sequences with the cDNA in ecotype Tsu-1 using MacVector software. Genetic diversity was analyzed as previously described [39]. Genomic sequences of the 29 alleles were aligned using ClustalX 2.0 [40]. The alignment results were exported into DnaSP 5.10.01 software [22]. The haplotype diversity, the nucleotide diversity, and the population mutation rate (Watterson estimator θw [41]) were estimated using the default parameters of the software in all cases [22].
To analyze the diversity differences of exons and introns, the exon and intron sequences were extracted from genomic sequences and analyzed as described above. The significance of the differences was tested with one-way ANOVA at 5% confidence level.
Acknowledgments
Many thanks to the reviewers and editors for their efforts in improving this paper, and Johan Meijer, Bioscience Center, Swedish University of Agricultural Sciences for providing some of the Arabidopsis ecotypes. This work was partly supported by the Natural Science Foundation of China (Grant Nos. 31070271, 31471561), and the Natural Science Foundation of Hainan Province (Grant No. 80669).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/17/2/262/s1.
Author Contributions
Jiaming Zhang conceived and designed the experiments; Lili Fu, Bingying Han, Deguan Tan, Meng Wang, and Mei Ding performed the experiments; Lili Fu analyzed the data; Jiaming Zhang and Lili Fu wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lombard V., Ramulu H.G., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZY) in 2013. Nucleic Acids Res. 2014;24 doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fahey J.W., Zalcmann A.T., Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry. 2001;56:5–51. doi: 10.1016/S0031-9422(00)00316-2. [DOI] [PubMed] [Google Scholar]
- 4.Barth C., Jander G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006;46:549–562. doi: 10.1111/j.1365-313X.2006.02716.x. [DOI] [PubMed] [Google Scholar]
- 5.Kim J.H., Jander G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 2007;49:1008–1019. doi: 10.1111/j.1365-313X.2006.03019.x. [DOI] [PubMed] [Google Scholar]
- 6.Xue J.P., Lenman M., Falk A., Rask L. The glucosinolate-degrading enzyme myrosinase in Brassicaceae is encoded by a gene family. Plant Mol. Biol. 1992;18:387–398. doi: 10.1007/BF00034965. [DOI] [PubMed] [Google Scholar]
- 7.Thangstad O.P., Winge P., Husebye H., Bones A. The myrosinase (thioglucoside glucohydrolase) gene family in Brassicaceae. Plant Mol. Biol. 1993;23:511–524. doi: 10.1007/BF00019299. [DOI] [PubMed] [Google Scholar]
- 8.Rask L., Andreasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 2000;42:93–113. doi: 10.1023/A:1006380021658. [DOI] [PubMed] [Google Scholar]
- 9.Husebye H., Chadchawan S., Winge P., Thangstad O.P., Bones A.M. Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 2002;128:1180–1188. doi: 10.1104/pp.010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson D., Chakrabarty R., Bejai S., Zhang J., Rask L., Meijer J. Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry. 2009;70:1345–1354. doi: 10.1016/j.phytochem.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., Andersson D., Meijer J. Characterisation of a new type of β-thioglucoside glycosidase (myrosinase), Arabidopsis thaliana; Proceedings of the VIII International Conference on Arabidopsis Research; Sevilla, Spain. 28 June–2 July 2002; pp. 6–25. [Google Scholar]
- 12.Wang M., Li D., Tan D., Gong S., Sun X., Meijer J., Zhang J. The two non-functional myrosinase genes TGG3 and TGG6 in Arabidopsis are expressed predominantly in pollen. Plant Sci. 2009;177:371–375. doi: 10.1016/j.plantsci.2009.06.003. [DOI] [Google Scholar]
- 13.Zhang J., Pontoppidan B., Xue J., Rask L., Meijer J. The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol. Plant. 2002;115:25–34. doi: 10.1034/j.1399-3054.2002.1150103.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang M., Li D., Sun X., Zhu Y.J., Nong H., Zhang J. Characterization of a root-specific β-thioglucoside glucohydrolase gene in Carica papaya and its recombinant protein expressed in Pichia pastoris. Plant Sci. 2009;177:716–723. doi: 10.1016/j.plantsci.2009.09.013. [DOI] [Google Scholar]
- 15.Nong H., Zhang J., Li D., Wang M., Sun X., Zhu Y.J., Meijer J., Wang H. Characterization of a novel β-thioglucosidase CpTGG1 in Carica papaya and its substrate-dependent and ascorbic acid-independent O-β-glucosidase activity. J. Integr. Plant Biol. 2010;52:879–890. doi: 10.1111/j.1744-7909.2010.00988.x. [DOI] [PubMed] [Google Scholar]
- 16.Xue J., Rask L. The unusual 5′ splicing border GC is used in myrosinase genes of the Brassicaceae. Plant Mol. Biol. 1995;29:167–171. doi: 10.1007/BF00019128. [DOI] [PubMed] [Google Scholar]
- 17.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 18.Burmeister W.P., Cottaz S., Rollin P., Vasella A., Henrissat B. High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base. J. Biol. Chem. 2000;275:39385–39393. doi: 10.1074/jbc.M006796200. [DOI] [PubMed] [Google Scholar]
- 19.Kleinwachter M., Selmar D. A novel approach for reliable activity determination of ascorbic acid depending myrosinases. J. Biochem. Biophys. Methods. 2004;59:253–265. doi: 10.1016/j.jbbm.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Shikita M., Fahey J.W., Golden T.R., Holtzclaw W.D., Talalay P. An unusual case of “uncompetitive activation” by ascorbic acid: Purification and kinetic properties of a myrosinase from Raphanus sativus seedlings. Biochem. J. 1999;341:725–732. doi: 10.1042/0264-6021:3410725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burmeister W.P., Cottaz S., Driguez H., Iori R., Palmieri S., Henrissat B. The crystal structures of Sinapis alba myrosinase and a covalent glycosyl–enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure. 1997;5:663–675. doi: 10.1016/S0969-2126(97)00221-9. [DOI] [PubMed] [Google Scholar]
- 22.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen L.M. Insecticide-resistant pollen beetles (Meligethes aeneus F) found in Danish oilseed rape (Brassica napus L) fields. Pest Manag. Sci. 2003;59:1057–1059. doi: 10.1002/ps.737. [DOI] [PubMed] [Google Scholar]
- 26.Kliebenstein D.J., Kroymann J., Mitchell-Olds T. The glucosinolate-myrosinase system in an ecological and evolutionary context. Curr. Opin. Plant Biol. 2005;8:264–271. doi: 10.1016/j.pbi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Bones A.M., Rossiter J.T. The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry. 2006;67:1053–1067. doi: 10.1016/j.phytochem.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Kusaba M., Dwyer K., Hendershot J., Vrebalov J., Nasrallah J.B., Nasrallah M.E. Self-incompatibility in the genus Arabidopsis: Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell. 2001;13:627–643. doi: 10.1105/tpc.13.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman-Broyles S., Boggs N., Farkas A., Liu P., Vrebalov J., Nasrallah M.E., Nasrallah J.B. S locus genes and the evolution of self-sertility in Arabidopsis thaliana. Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang C., Toomajian C., Sherman-Broyles S., Plagnol V., Guo Y.-L., Hu T.T., Clark R.M., Nasrallah J.B., Weigel D., Nordborg M. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 31.Yang L., Gaut B.S. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol. Biol. Evol. 2011;28:2359–2369. doi: 10.1093/molbev/msr058. [DOI] [PubMed] [Google Scholar]
- 32.Wright S.I., Lauga B., Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- 33.Charlesworth B. Evolutionary rates in partionally self-fertilizing species. Am. Nat. 1992;140:126–148. doi: 10.1086/285406. [DOI] [PubMed] [Google Scholar]
- 34.Zhang P.G., Huang S.Z., Pin A.-L., Adams K.L. Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis. Mol. Biol. Evol. 2010;27:1686–1697. doi: 10.1093/molbev/msq054. [DOI] [PubMed] [Google Scholar]
- 35.Barbazuk W.B., Fu Y., McGinnis K.M. Genome-wide analyses of alternative splicing in plants: Opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y.W., Lai K.N., Tai P.Y., Ma D.P., Li W.H. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 1999;48:597–604. doi: 10.1007/PL00006502. [DOI] [PubMed] [Google Scholar]
- 37.Bechtold N., Ellis J., Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- 38.Jefferson R.A., Kavanagh T.A., Bevan M.W. Gus fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y., Ma S., Huang M., Peng M., Bog M., Sree K.S., Appenroth K., Zhang J. Species distribution, genetic diversity and barcoding in the duckweed family (Lemnaceae) Hydrobiologia. 2015;743:75–87. doi: 10.1007/s10750-014-2014-2. [DOI] [Google Scholar]
- 40.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.