Fig. 6.
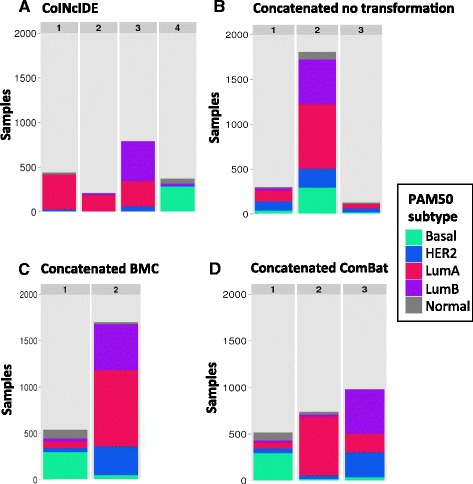
CoINcIDE versus concatenated results. Bar plots summarizing the counts for each PAM50 centroid subtype for each subtyping result from (a) PAM50 restricted 35-gene set CoINcIDE k-mean clustering, (b) concatenated matrix with no transformation, (c) concatenated matrix with gene-wise batch mean centering (BMC), and (d) concatenated matrix with ComBat. All analyses used k-means consensus clustering using the Proportion of Ambiguous Clusters (PAC) to select the number of clusters. These bar plots used only the 35-gene set to assign patients to PAM50 centroid subtypes (Additional file 1: Table S1). PAM50 centroid assignments were made on the pre-transformed concatenated datasets to allow for direct comparison between all four subtype discovery methods. See Additional file 3: Figure S9B for the corresponding CoINcIDE network figure for (a)
