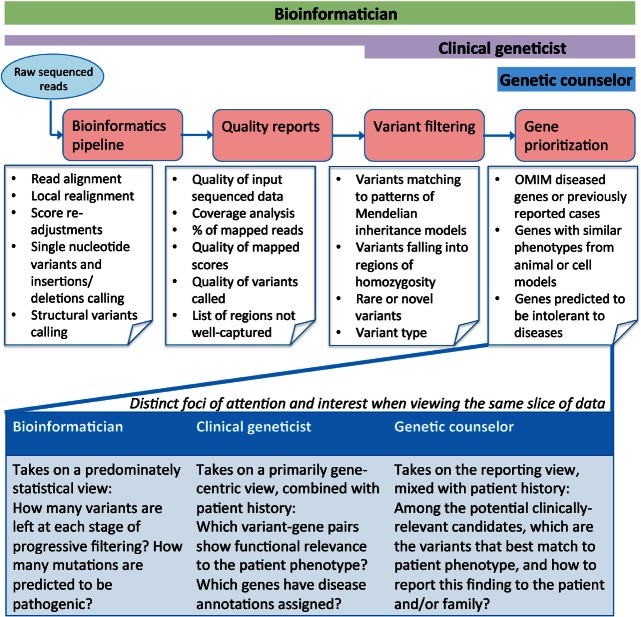
Figure 1:
Beginning with raw sequenced reads, the exome analysis pipeline can be conceptualized into 4 distinct compartments: generation of alignments and variant calls, assessment of data quality, filtering of variants based on genetic models, and prioritization of genes based upon biological functions. The details of the components are annotated largely in the context of genetic diagnosis for rare/complex disorders (refer to Supplementary S12 for discussion on other clinical uses of genomic data). The bars above represent the intensity of user engagement at each step. Bioinformaticians preferred to be involved in every step, with equal attention devoted to all compartments. Clinical geneticist, despite placing heavier emphasis on the final 2 stages, indicated they would ideally like to be involved in every step too, but they faced difficulties in carrying out the first and second steps (e.g., pipeline execution and quality assessment), which may be attributed to software usability. Genetic counselors (and general physicians, not shown) indicated they would focus on the final output of candidate variants, to which they could apply their domain knowledge to select clinically relevant genes. The text in the lower portion of the figure highlights how the same step in the informatics pipeline (e.g., variant data) can be viewed differently across domain experts.