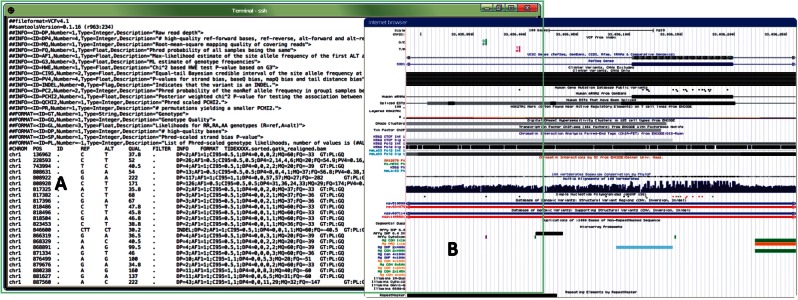
Figure 4:
A graphical representation of the key features desired by bioinformaticians when visualizing/filtering variant sets. (A) Analyzing variants within a terminal environment by informaticians allows manipulation of the variant files via custom scripts and/or external command-line programs. (B) Variants are preferred to be visualized within a genome browser (e.g., University of California, Santa Cruz (UCSC) Genome Browser73) where genomic neighborhood landmarks and any additional relevant biological information (e.g., SNPs, conservation) can be displayed alongside.