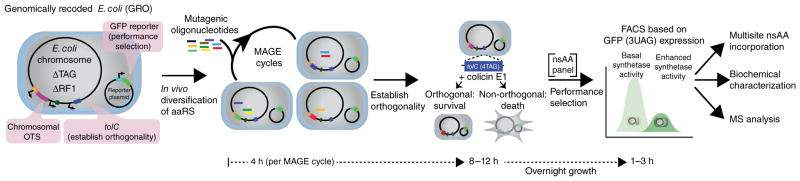
Figure 2.
Evolution of chromosomally integrated aaRS variants. The genomically recoded organism (GRO) is engineered to contain a single chromosomal copy of the aaRS for diversification using MAGE, a negative-selection marker for removal of nonorthogonal translation systems (OTS) (capable of incorporation of natural amino acids), and a GFP marker for fluorescence-based identification and isolation of improved variants. Site-directed mutagenesis of chromosomally integrated translation components by MAGE generates a highly diversified population, which is subsequently subjected to tolC- and colicin E1–mediated negative selection in the absence of nsAAs. UAG suppression in GFP(3UAG) enables FACS of orthogonal aaRS libraries in the presence of the desired nsAA to identify improved variants. The selected aaRS variants are evaluated for multi-site nsAA incorporation, in vitro activity and protein purity.