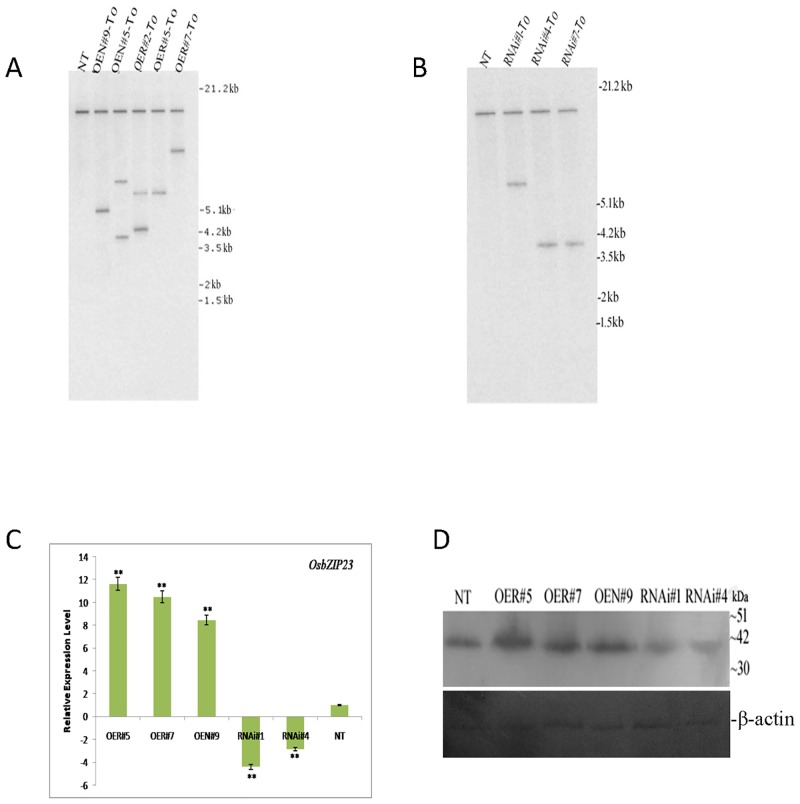
Fig 5. Molecular analyses of transgenic and NT plants for studying the CDS activity of OsbZIP23 gene.
(A) Southern blot analysis of T0 transformants of OEN and OER developed with the genetic construct of OsbZIP23 CDS from O. rufipogon (S3 Fig) and O. nivara (S4 Fig). The genomic DNA was digested with HindIII and probed with the 739 bp OsbZIP23 CDS. (B) Southern blot analysis of T0 transformants of RNAi developed with gene silencing construct (S5 Fig) using HindIII digested genomic DNA and probed with 739 bp OsbZIP23 CDS. Lane NT- non-transgenic, Lane M- molecular weight marker. (C) Real time PCR analysis showing relative expression level of OsbZIP23 in three OE lines, two RNAi lines and non-transgenic (NT) plants in vegetative stage, where rice polyubiquitin1 (OsUbi1) gene was taken as internal reference. Data bars represent the mean ±SD of triplicate measurement. Statistical analysis of Student’s t test indicated significant differences (* P<0.05, **P<0.01). (D) Western blot analysis showing the expression level of OsbZIP23 protein in three OE lines, two RNAi lines and NT plant (upper panel). Equal loading of protein was confirmed by using β-actin antibody (lower panel).