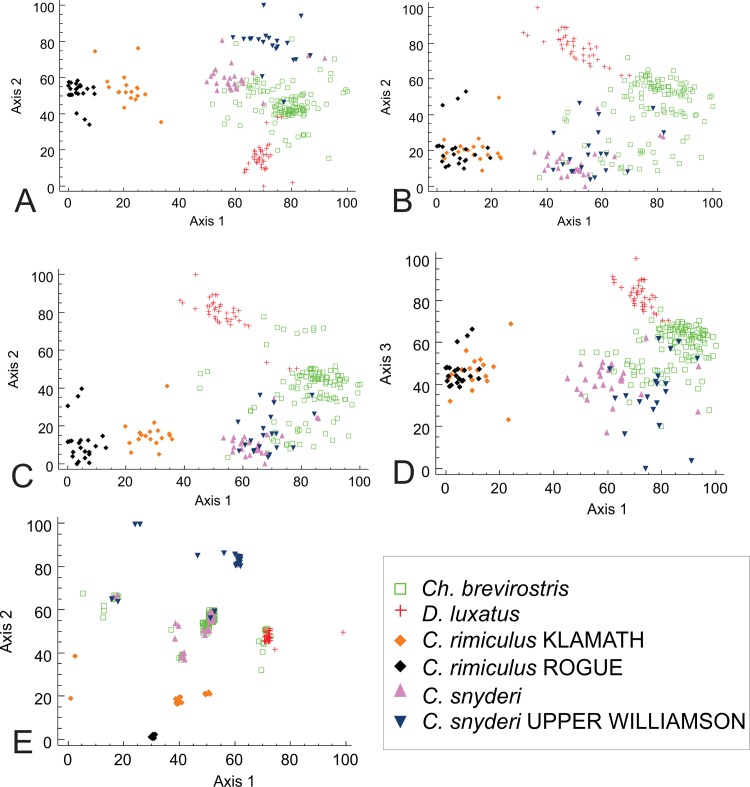
Fig 7. Two major axes for solutions from nonmetric multidimensional scaling of Klamath and Rogue basin suckers.
A-D were rotated to align with gillraker counts, E rotated to align with the LUX haplotype. All axes scaled to percentages. A. Two dimensional solution (k = 2) using all morphological and genetic characters. B. Two dimensional solution (k = 2) using morphological data only. C. Two dimensional solution (k = 2) using morphological and mtDNA. D. First and third axes of three dimensional solution (k = 3) using morphological and nuclear DNA. E. Two dimensional solution (k = 2) using mitochondrial and nuclear data with points jittered to reduce overlap.