Figure 4.
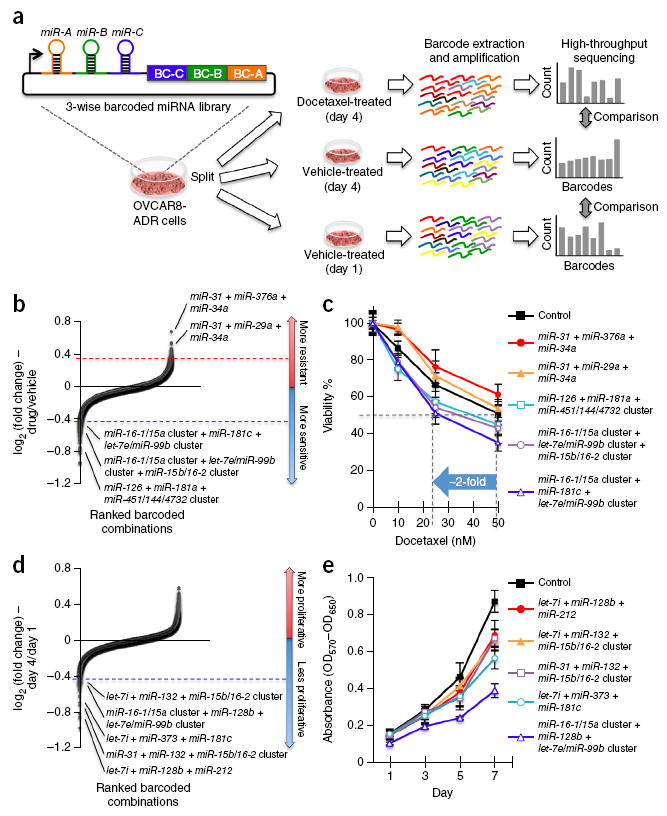
Three-wise combinatorial screens identify miRNA combinations modifying docetaxel sensitivity or proliferation in cancer cells. (a) OVCAR8-ADR cells infected with the three-wise combinatorial miRNA library were split into three groups, and treated with 25 nM of docetaxel or vehicle for 4 days, or cultured with vehicle for 1 day. The barcodes of each combinatorial miRNA construct were amplified by PCR from the genomic DNA within the cell pools in an unbiased fashion, and counted using Illumina HiSeq. (b–e) Three-wise miRNA combinations and validation. Combinations that modulated docetaxel sensitivity (b) and proliferation (d) of OVCAR8-ADR cells were ranked by their mean log2 ratios from two biological replicates. The log2 ratios of the normalized barcode count for docetaxel (25 nM)-treated versus 4-day vehicle-treated cells (b) and 4-day versus 1-day cultured cells (d) were used as the measure of docetaxel sensitivity and cell proliferation, respectively. For validation of combinations, OVCAR8-ADR cells infected with combinations that altered docetaxel sensitivity were treated with docetaxel at different doses for 3 days (c) and those that modulated proliferation were treated for the indicated time periods (e). Cell viability was measured by the MTT assay, and was compared to the no-drug control (c; n ≥ 5) or characterized by absorbance measurements (OD570−OD650) (e; n ≥ 4). Data represent mean ± s.d.
