Abstract
Abstract
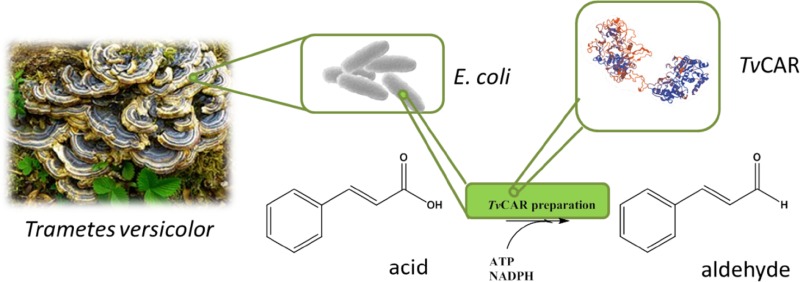
The first carboxylate reductase from Trametes versicolor was identified, cloned, and expressed in Escherichia coli. The enzyme reduces aromatic acids such as benzoic acid and derivatives, cinnamic acid, and 3-phenylpropanoic acid, but also aliphatic acids such as octanoic acid are reduced.
Graphical abstract
Keywords: Bioreduction, Carboxylate reductase, Carboxylic acids, Aldehydes, Enzymes
Introduction
The reduction of carboxylic acids is a challenge both for chemical transformations and biotransformations. From the thermodynamic viewpoint, the carboxylic acid moiety is in an energetically favored state, shows little reactivity and needs a high level of activation to participate in chemical reactions. Due to the high stability of carboxylates, examples for their direct reduction are rare and usually an initial activation step (e.g., to the carboxylic ester or to the acyl halide) is required. The reduction of the COOH group or its derivatives yields the respective aldehyde in the first step; because aldehyde is more reactive than the carboxylic acid, the aldehyde does not accumulate but is typically reduced further to the respective primary alcohol. Certain reaction conditions may promote the accumulation of aldehydes instead of alcohol: the well-known examples are the reduction of acid chloride with lithium tri-t-butoxy aluminum hydride [1] or DIBALH [2]. In the past two decades, several selective chemical reductions to alcohols or aldehydes using hydrosilanes have also been described [3]. In laboratory practice, however, the acid is often fully reduced to the alcohol and then re-oxidized to the aldehyde [4], e.g., via the classical Swern or Dess–Martin oxidation [5]. Other oxidation protocols use chromate or manganese reagents, but also a large variety of methods using molecular oxygen in combination with immobilized transition metal catalysts have recently been developed [6]. For example, ruthenium or platinum on carbon in organic solvents serves as the catalyst for the oxidation of aromatic alcohols to the corresponding aldehydes [7, 8]. The use of organic solvents, toxic reagents, undesired reaction conditions or the high demand of reducing equivalents, however, induces a strong wish for one step alternatives to the selective preparation of aldehydes from acids.
A green substitution to chemical methods is the use of biocatalysts with carboxylic acid reduction ability. The first biocatalyzed reduction of carboxylic acid substrates was accomplished by the white-rot fungus Trametes versicolor [9]. A significant number of subsequent reports followed that described the reduction of carboxylic acids also by other fungi such as Fomes fomentarius [10], Pycnoporus cinnabarinus [11], Aspergillus sp. [11, 12], Bjerkandera sp. [13], Mucor sp. [14], and many others [15, 16]. However, to date, the amino acid sequence of only one single fungal carboxylate reductase has been elucidated from Aspergillus terreus [17, 18]. Herein, the sequence and substrate scope of the first carboxylate reductase from T. versicolor have been explored.
Results and discussion
Trametes versicolor (alias: Polystictus versicolor) was reported to reduce benzoic acid derivatives to mixtures of aldehydes and alcohols [9]. The enzyme responsible for the first reduction step from the acid to the aldehyde had not been identified to this date, nor has a particular enzyme been purified and characterized. To discover the enzymatic activity on sequence level, the fungal carboxylate reductase enzyme sequence and three sequences of homologous bacterial ATP and NADPH-dependent carboxylate reductases were used as templates to search within the non-redundant protein sequences on NCBI with the restriction to the organism T. versicolor. The hits from this search were all annotated as acetyl-CoA synthetase-like proteins. Aspergillus terreus ATEG_03630 [17] is classified as CaiC for the adenylation domain and Thioester-redct for the reduction domain by the multi-domain model, whereas the CARs from Nocardia iowensis [19], Mycobacterium marinum [20], and Segniliparus rotundus [21] are classified as FAA1 and Lys2B. From the most significant hits of the search, one sequence was classified to consist of an FAA1 adenylation domain in combination with a Thioester-reductase domain. Therefore, it seemed most likely that this sequence would display carboxylate reductase activity.
Since homologous bacterial carboxylate reductases were shown to require the attachment of a phosphopantethein moiety [22], the protein was expressed from the pETDUET1 vector that harbored simultaneously Escherichia coli phosphopantethein transferase for post-translational modification of the carboxylate reductase and the coding sequence of the putative CAR enzyme with an N-terminal fusion tag. Despite codon optimization of the fungal gene sequence for expression in E. coli, the amount of soluble protein was disappointingly low and the majority of the protein was found in the insoluble fraction. The protein was purified via affinity chromatography, yielding T. versicolor CAR (TvCAR) in enriched form. As known CAR enzymes have been well expressed heterologously in E. coli in their apo-form [22], the reason is unlikely insufficient post-translational phosphopantetheinylation. Optimization of the expression conditions, such as the choice of a weaker promoter, the use of chaperones [23], or ultimately a fungal expression system, will likely result in significant improvements with respect to enzyme yield.
In comparison to published CAR enzymes, T. versicolor CAR is most similar to Aspergillus terreus CAR, albeit with only 24 % identity according to UniProtAlign, whereas N. iowensis CAR and M. marinum CAR exhibit 17 % and S. rotundus CAR 16 % identity, respectively.
Carboxylate reductase enzymes rely on ATP and NADPH as cosubstrates. As typical for adenylating enzymes, the presence of Mg2+ is important due to its interaction with ATP [24]. TvCAR preparation was assayed using the conditions as described in the experimental section. In addition, control reactions without the addition of substrate (E)-cinnamic acid (1a), ATP, or MgCl2 were carried out. NADPH was not oxidized in the absence of ATP or substrate. When the reaction was carried out in TrisHCl buffer without MgCl2, the carboxylate reduction rate was diminished by approximately 30 %. It needs to be noted that the enzyme preparation contained 10 mM MgCl2, resulting in 0.5 mM MgCl2 end concentration in this particular reaction.
TvCAR was subjected to an NADPH depletion assay in the presence of different carboxylic acids. Enzymatic activity was observed for the substrates listed in Table 1. In the presence of, e.g. phenylacetic acid, mandelic acid, 2-pyrrole-, 2-pyridine-, 3-indole-, and 2-pyrazinecarboxylic acid as well as short chain aliphatic acids levulinic acid, pentanoic acid and hexanoic acid, NADPH was not oxidized by TvCAR. The fact that 1a, benzoic acid (2a), 3-hydroxy- and 3-methoxybenzoic acid (3a, 4a) as well as 4-methoxybenzoic acid (5a) are reduced, but phenylacetic acid is not, is consistent with the observations of Farmer et al. [9]. However, 2-hydroxy- and 2-methoxybenzoic acids were not reduced by TvCAR under the conditions used. This supports the idea that T. versicolor harbors more than one CAR enzyme and CAR (XP_008043822.1) is partly but not fully responsible for the reactions observed in 1959. As an analog of 1a, 3-phenylpropanoic acid (6a) was also reduced. Surprisingly, also aliphatic carboxylates from C7 to C9 (7a–9a) were converted, whereas short chain acids and longer chain acids were not accepted as substrates under these conditions (Table 1).
Table 1.
Substrate scope of Trametes versicolor carboxylate reductase
| Entry | Substrate | Specific activitya/mU mg−1 |
|---|---|---|
| 1 | (E)-Cinnamic acid (1a) | 41 ± 4 |
| 2 | Benzoic acid (2a) | 30 ± 9 |
| 3 | 3-Hydroxybenzoic acid (3a) | 22 ± 8 |
| 4 | 3-Methoxybenzoic acid (4a) | 23 ± 5 |
| 5 | 4-Methoxybenzoic acid (5a) | 33 ± 2 |
| 6 | 3-Phenylpropanoic acid (6a) | 34 ± 7 |
| 7 | Heptanoic acid (7a) | 34 ± 7 |
| 8 | Octanoic acid (8a) | 35 ± 4 |
| 9 | Nonanoic acid (9a) | 33 ± 1 |
aOne activity unit is defined as the amount of enzyme preparation catalyzing the oxidation of 1 µM of NADPH per minute
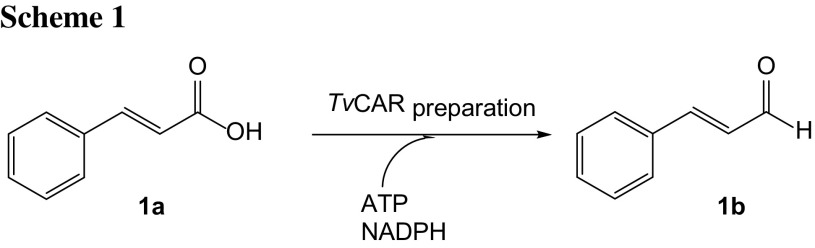
To investigate the identity of the reduction product, (E)-cinnamic acid (1a) was subjected to a biotransformation reaction in the presence of the TvCAR enzyme and excess of co-factors. The reaction was analyzed after extraction and derivatization of the remaining substrate 1a by GC/MS analysis, revealing cinnamaldehyde (1b) as the sole product in a mixture with methyl cinnamate (1d) (Scheme 1).
Conclusion
The currently available portfolio of carboxylate reductase enzymes is limited to a handful of protein sequences. With the aim to enlighten new biocatalysts, we identified a putative acetyl-CoA synthetase-like protein from T. versicolor, expressed it in E. coli and confirmed that this protein indeed shows carboxylate reductase activity. The enzyme exhibits a promising substrate scope; however, its heterologous expression is a clear limitation at the moment.
Experimental
Substrates, product references, DTT, and trimethylsilyldiazomethane (solution, 2 M in diethyl ether) were purchased from Sigma Aldrich and used without further purification. ATP was obtained from Roche Diagnostics. NADPH and MES were purchased from Roth, IPTG from Serva, and MgCl2 from Merck.
Cloning and expression
Four literature known carboxylate reductase enzyme sequences (Q6RKB1.1, WP_012393886.1, WP_013138593.1, and XP_001212808.1) were used as search templates for a blastp search against non-redundant protein sequences with the restriction to the organism T. versicolor (taxid:5325). A gene, coding for the protein sequence of NCBI accession code XP_008043822.1 was ordered as synthetic gene with optimization of the genetic code for expression in E. coli. The protein was expressed from the pETDUET1 vector with the E. coli pptase (NCBI accession code CAQ31055.1) cloned into the first multiple cloning site and N-terminally HIS-tagged CAR sequences in the second multiple cloning site. The plasmid was transfected to E. coli BL32(DE3) Star and colonies selected on LB/Amp. The enzyme was expressed using standard conditions. The cells were harvested by centrifugation and stored at −20 °C. After thawing, the cells were disrupted by sonication and the protein purified by nickel affinity chromatography, using the gravity flow protocol. The protein containing fractions were pooled and dialyzed overnight at 4 °C into 50 mM MES buffer, pH 7.5, containing 10 mM MgCl2, 1 mM EDTA, and 1 mM DTT. Aliquots of the resultant, slightly turbid protein solution were shock frozen in liquid nitrogen and stored at −80 °C.
Substrate screening
The capability of the protein to reduce carboxylic acids was determined using a photometric NADPH depletion assay. Therefore, potential carboxylic acid substrates were dissolved in KOH (0.1 M in water) or DMSO. The assay composition was as follows: The substrates (10 mm3 of 100 mM stock solution) were added to 160 mm3 of TrisHCl buffer (100 mM, pH 7.5, containing 10 mM MgCl2). Subsequently, 10 mm3 of NADPH (10 mM in water), 10 mm3 of ATP (20 mM in water), and 10 mm3 of enzyme preparation (3.85 mg/cm3 with an estimated purity of 50 %) were added. Each reaction was measured four times in parallel in UV-Star 96 well plates (Greiner). The depletion of NADPH was followed on a Synergy Mx Platereader at 340 nm and 28 °C for 10 min. Blank reactions without enzyme preparation were carried out in parallel.
Biotransformation
1a (10 mm3 of 100 mM stock in DMSO) was added to 120 mm3 of MES buffer (50 mM, pH 7.5, containing 10 mM MgCl2, 1 mM EDTA, and 1 mM DTT). Subsequently, 10 mm3 of NADPH (100 mM in water), 50 mm3 of ATP (20 mM in water), and 60 mm3 of CAR enzyme preparation (4.5 mg/cm3 total protein, containing approximately 50 % of CAR) were added. The reaction proceeded in a Thermomixer at 28 °C and 300 rpm. After 2 h and overnight incubation, the reaction was acidified by the addition of aqueous HCl (4 N, 50 mm3) and the products were extracted with ethyl acetate (2 × 0.3 cm3). The extracts were dried over Na2SO4 and TMS-diazomethane solution (2 mm3, 2 M in diethyl ether) was added for the derivatization of 1a to the corresponding methyl ester for GC analysis. GC–MS analyses were performed on an Agilent 7890A Series GC system equipped with a 5975C MS system, using a (5 % phenyl) methylpolysiloxane capillary column (HP-5MS, 30 m, 0.25, 0.25 µm film) with He as the carrier gas. The method was 100 °C hold 0.5 min, 10 °C/min to 300 °C. Retention times were: 1b: 6.10 min; cinnamic alcohol (1c, a potential product derived from over-reduction): 6.47 min; methyl cinnamate (1d): 7.44 min.
Acknowledgments
Open access funding provided by Graz University of Technology. The Austrian science fund is kindly acknowledged for financial support (Elise-Richter fellowship V415-B21).
References
- 1.Brown HC, Rao BCS. J Am Chem Soc. 1958;80:5377. doi: 10.1021/ja01553a014. [DOI] [Google Scholar]
- 2.Chandrasekhar S, Kumar MS, Muralidhar B. Tetrahedron Lett. 1998;39:909. doi: 10.1016/S0040-4039(97)10688-8. [DOI] [Google Scholar]
- 3.Addis D, Das S, Junge K, Beller M. Angew Chem Int Ed. 2011;50:6004. doi: 10.1002/anie.201100145. [DOI] [PubMed] [Google Scholar]
- 4.Kotha S, Ravikumar O (2015) Beilstein J Org Chem 11:1259 [DOI] [PMC free article] [PubMed]
- 5.Dess DB, Martin JC. J Org Chem. 1983;48:4155. doi: 10.1021/jo00170a070. [DOI] [Google Scholar]
- 6.Bäckvall J-E (2004) Modern oxidation methods. Wiley-VCH Verlag GmbH and Co. KGaA, Weinheim
- 7.Mori S, Takubo M, Makida K, Yanase T, Aoyagi S, Maegawa T, Monguchi Y, Sajiki H. Chem Commun. 2009;34:5159. doi: 10.1039/b908451g. [DOI] [PubMed] [Google Scholar]
- 8.Korovchenko P, Donze C, Gallezot P, Besson M. Catal Today. 2007;121:13. doi: 10.1016/j.cattod.2006.11.007. [DOI] [Google Scholar]
- 9.Farmer VC, Henderson MEK, Russell JD. Biochim Biophys Acta. 1959;35:202. doi: 10.1016/0006-3002(59)90349-X. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa H, Schubert WJ, Nord FF. Arch Biochem Biophys. 1963;100:140. doi: 10.1016/0003-9861(63)90044-4. [DOI] [PubMed] [Google Scholar]
- 11.Arfmann HA, Abraham WR. Z Naturforsch C. 1993;48:52. doi: 10.1515/znc-1993-1-210. [DOI] [PubMed] [Google Scholar]
- 12.Motedayen N, Ismail MBT, Nazarpour F. African J Biotechnol. 2013;12:6618. doi: 10.5897/AJB2013.12416. [DOI] [Google Scholar]
- 13.Lauritsen FR, Lunding A. Enzyme Microb Technol. 1998;22:459. doi: 10.1016/S0141-0229(97)00237-8. [DOI] [Google Scholar]
- 14.Ma L, Liu X, Liang J, Zhang Z. World J Microbiol Biotechnol. 2011;27:2133. doi: 10.1007/s11274-011-0677-7. [DOI] [Google Scholar]
- 15.Napora-Wijata K, Strohmeier GA, Winkler M. Biotechnol J. 2014;9:822. doi: 10.1002/biot.201400012. [DOI] [PubMed] [Google Scholar]
- 16.da Silva Amaral L, Rodrigues-Filho E (2015) J Mol Catal B Enzym 113:90
- 17.Wang M, Zhao H. ACS Catal. 2014;4:1219. doi: 10.1021/cs500039v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Beissner M, Zhao H. Chem Biol. 2014;21:257. doi: 10.1016/j.chembiol.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He A, Li T, Daniels L, Fotheringham I, Rosazza JPN. Appl Environ Microbiol. 2004;70:1874. doi: 10.1128/AEM.70.3.1874-1881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhtar MK, Turner NJ, Jones PR. Proc Natl Acad Sci. 2013;110:87. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan Y, Yao P, Chen X, Liu X, Zhang R, Feng J, Wu Q, Zhu D. J Mol Catal B Enzym. 2015;115:1. doi: 10.1016/j.molcatb.2015.01.014. [DOI] [Google Scholar]
- 22.Venkitasubramanian P, Daniels L, Rosazza JPN. J Biol Chem. 2007;282:478. doi: 10.1074/jbc.M607980200. [DOI] [PubMed] [Google Scholar]
- 23.Weis R, Winkler M, Schittmayer M, Kambourakis S, Vink M, Rozzell JD, GLieder A (2009) Adv Synth Catal 351:2140
- 24.Schmelz S, Naismith JH. Curr Opin Struct Biol. 2009;19:666. doi: 10.1016/j.sbi.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]