Figure 3.
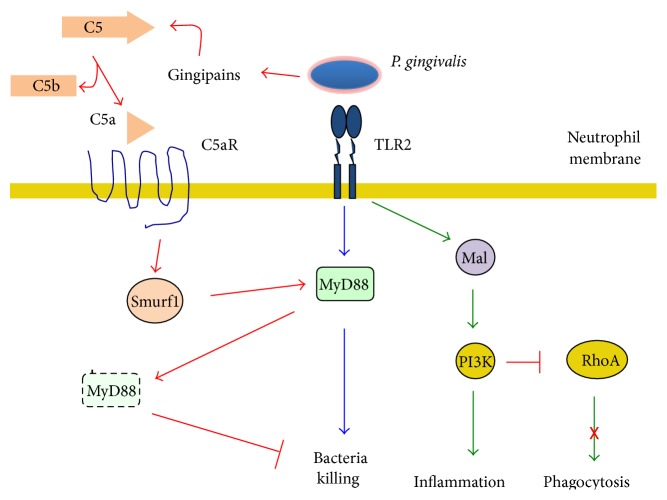
P. gingivalis can manipulate both complement and TLR signaling to induce bacterial persistence. The bacterium P. gingivalis is recognized by the TLR2 on neutrophils and activates the adaptor protein MyD88 for bacterial elimination (blue arrows). However, P. gingivalis produce gingipains, enzymes that function as complement C5 convertases. C5a complement fragment binds to its receptor (C5aR), which induces the E3 ligase Smurf1 to ubiquitinate MyD88 and mark if for proteosomal degradation (red arrows), effectively inhibiting bacteria killing. Interestingly, binding of P. gingivalis to the TLR2 also induces inflammation through a signaling pathway involving the adaptor molecule Mal and the enzyme phosphatidylinositol 3-kinase (PI-3K) (green arrows). Also, the same pathway can prevent actin polymerization through inhibition of the small GTPase RhoA and block phagocytosis.