Figure 5.
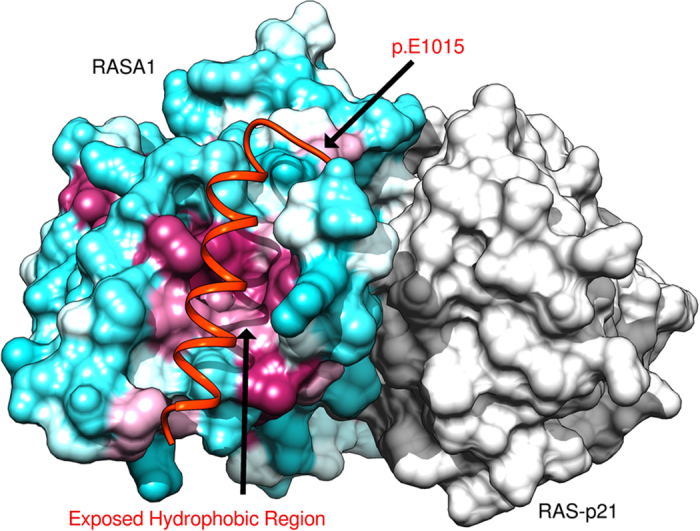
Graphical representation of RASA1 Ras-Gap domain and Ras-p21 crystal structure detailing the impact of the identified p.E1015* (c.3043G>T) mutation. The surface of RASA1 Ras-Gap domain (residues 718–1037) surface is colored based on the Kyte–Doolittle hydrophobicity scale (hydrophobic regions in purple and hydrophilic areas in turquoise) and the Ras-p21 is in shown with gray surface. The p.E1015* variant would remove the C-terminal helix (orange) and would act to destabilize the GTPase-activating protein (GAP) domain by exposing the hydrophobic region to solvent. Images generated with chimera.14
