Table 1. Data Collection and Refinement Statistics.
| INPP5B | ||
|---|---|---|
| BiPh(3,3′,4,4′,5,5′)P6 | Bz(1,2,4,5)P4 | |
| PDB ID | 5A7I | 5A7J |
| Data Collection | ||
| synchrotron | BESSY | BESSY |
| beamline | BL14.1 | BL14.1 |
| wavelength (Å) | 0.91841 | 0.91841 |
| resolution range (Å) | 34.29–2.89 | 48.48–2.90 |
| (outest shell) | (3.05–2.89) | (3.06–2.9) |
| space group | P41212 | P21 |
| unit-cell dimensions (Å) | a = 96.97 | a = 52.26 |
| b = 96.97 | b = 94.44 | |
| c = 152.14 | c = 78.31 | |
| unit-cell angles (deg) | α = β = γ = 90 | α = γ = 90, β = 106.32 |
| completeness (%) | 99.9 (100.0) | 99.7 (99.7) |
| unique reflections | 16922 | 16223 |
| mean (I)/SD (I)a | 21.2 (2.2) | 6.9 (2.3) |
| redundancy | 7.0 (7.2) | 3.6 (3.6) |
| Rmeas (%)b | 7.5 (90.7) | 19.4 (67.9) |
| Refinement | ||
| resolution range (Å) | 34.29–2.89 | 48.48–2.90 |
| (outest shell) | (2.97–2.89) | (3.10–2.90) |
| Rcryst (%)c | 19.8 (33.8) | 20.2 (21.9) |
| Rfree (%)d | 22.3 (34.5) | 25.0 (31.0) |
| Model Content | ||
| protein atoms | 2545 | 4971 |
| ligand atoms | 42 | 52 |
| metal atoms | 1 | 0 |
| water molecules | 28 | 76 |
| others | 16 | 26 |
| average B factors (Å2) | ||
| protein atoms | 83.59 | 39.10 |
| ligand atoms | 76.44 | 65.84 |
| metal atoms | 66.37 | n.d. |
| water molecules | 71.94 | 19.15 |
| other atoms | 111.59 | 63.89 |
| r.m.s.d. bonds (Å) | 0.007 | 0.008 |
| r.m.s.d. angles (deg) | 1.191 | 1.000 |
| Ramachandran plot (%)e | ||
| (favored, outliers) | 95.82, 0.32 | 94.95, 0 |
Mean (I)/SD is the mean ratio for all reflections of ⟨Ih⟩/SD ⟨Ih⟩, where for each unique reflection h, ⟨Ih⟩ is the weighted mean of measured Ih, and SD ⟨Ih⟩ is the mean of estimated error SD(I).
Rmeas = 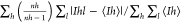
Rcryst = 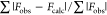 , where Fobs and Fcalc are the observed and calculated
structure factors, respectively.
, where Fobs and Fcalc are the observed and calculated
structure factors, respectively.
Rfree is the same as Rcryst but based on a subset of 5% (5.1% and 5.0% for INPP5B-BiPh(3,3′,4,4′,5,5′)P6 and INPP5B-Bz(1,2,4,5)P4, respectively) of reflections omitted during refinement.
Values were computed by Molprobity (ref (26)).
