Figure 1.
Smc ATPase Activity Determines the Chromosomal Distribution of Smc/ScpAB
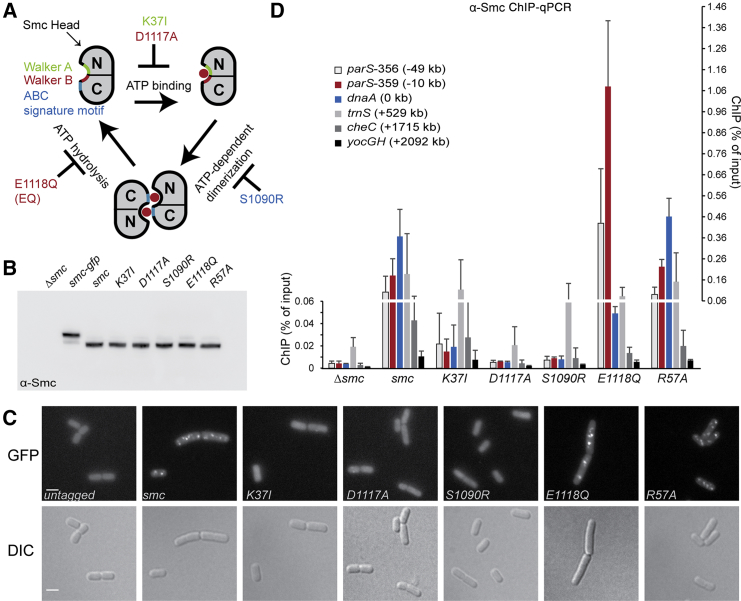
(A) Schematic representation of the Smc ATPase cycle.
(B) Immunoblotting of ATPase mutant Smc proteins with α-Smc antiserum. Whole-cell extracts from strains BSG1002, BSG1007–BSG1008, BSG1067, BSG1045–BSG1047, and BSG1083. See also Figure S1B.
(C) Fluorescence images of cells harboring mGFP-tagged Smc alleles: BSG1002, BSG1067–BSG1068, BSG1855–BSG1857, and BSG1881. Scale bar, 2 μm. Differential interference contrast (DIC) (bottom) and GFP fluorescence images (top) are shown. Quantification of foci number per cell is given in Figure S1H.
(D) ChIP-qPCR analysis of cells from strains BSG1002, BSG1007–BSG1008, BSG1045–BSG1047, and BSG1083 using α-Smc antiserum. Error bars were calculated from two independent experiments as SD. Please note that values of ChIP enrichment below and above 0.06% are displayed on different scales given on the left and right side of the graph, respectively. The analysis of chromosomal loci harboring highly transcribed genes (such as the tRNA cluster trnS) generally produce ambiguous results with relatively high levels of ChIP signal in control samples (Δsmc). This seems to be a widely observed phenomenon in ChIP and it remains unclear whether the enrichment is physiologically relevant.
See also Figure S1.