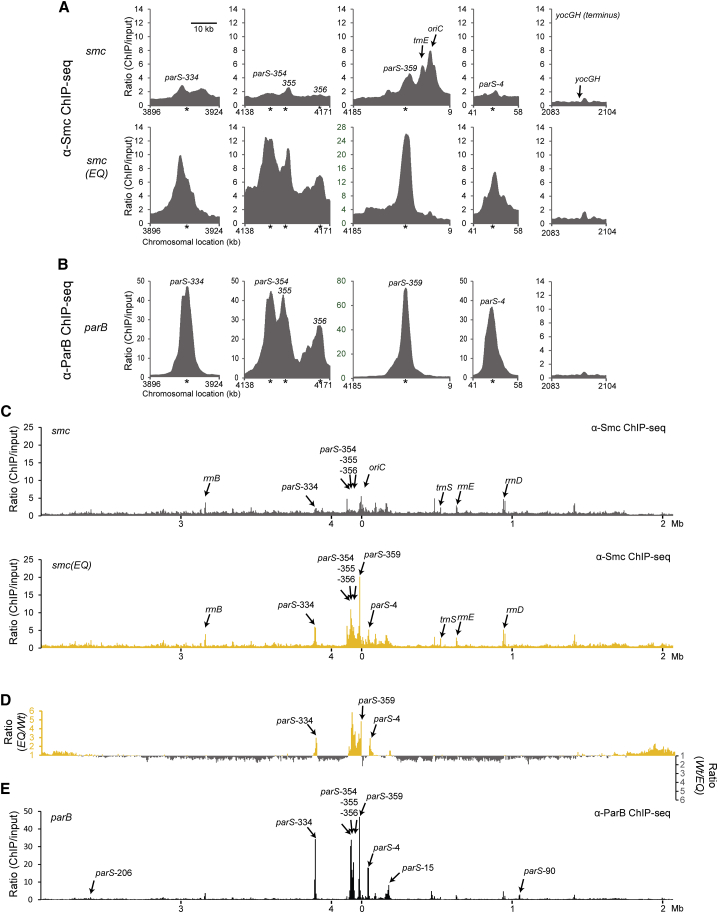
Figure 2.
Hydrolysis Mutant but Not Wild-Type Smc Co-localizes with ParB/parS
(A) Close-up view of ChIP-seq profiles for wild-type Smc (BSG1002) (top panel) and Smc(EQ) (BSG1008) (bottom panel) generated using antiserum raised against the Bs Smc protein. Sequence reads were mapped to 1 kb windows spaced at 100-bp intervals and normalized for input DNA as follows. The number of reads for the ChIP sample in a given window was divided by the number of reads in the input sample for the same window (after normalizing the total number of reads). Raw input and ChIP data are shown in Figure S2. Axes labeled in green color highlights different scaling. Asterisks indicate the positions of parS sites.
(B) Close-up view of the ChIP-seq profile of ParB protein (from BSG1470 cells) generated using antiserum raised against purified BsParB-His6 protein. Data analysis and presentation as in (A).
(C) Whole-genome views of data presented in (A). Sequence reads were mapped to 5-kb windows spaced at 5-kb intervals across the genome and normalized for input DNA.
(D) To highlight differences between the distribution of wild-type Smc and Smc(EQ) the normalized ratios for Smc(EQ) in a given window was divided by the equivalent ratio for Smc(wt). Numbers above one are shown in yellow colors (axis on the left side). For numbers below one, the inverse ratio was calculated and displayed in gray colors (axis on the right side).
(E) Whole-genome view of the ParB ChIP-seq data presented in (B). Data analysis as in (C).
See also Figure S2.