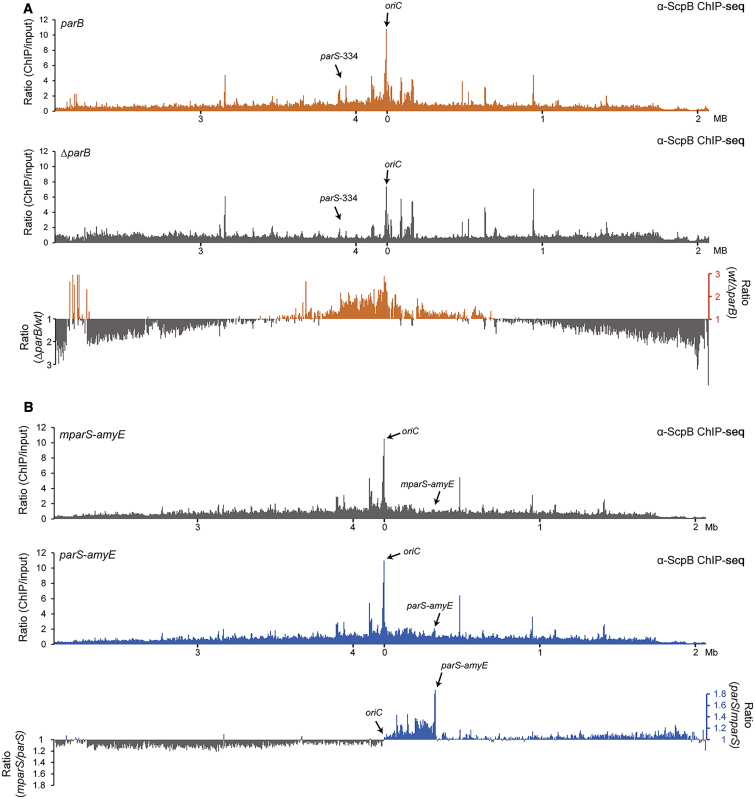
Figure 6.
Smc/ScpAB Relocates from parS Loading Sites to Distant Parts of the Chromosome upon ATP Hydrolysis
(A) ChIP-seq using α-ScpB antiserum on strains BSG1002 (parB) (top panel) and BSG1052 (ΔparB) (middle panel). Reads were mapped to 5-kb bins. Signals for IP samples were divided by the signals of the normalized input. Ratios were calculated by dividing the values obtained for the wild-type strain by the numbers of the ΔparB strain (bottom panel). All values above one are shown in orange colors. For all other windows the inverse ratio was calculated and displayed in gray colors.
(B) ChIP-seq using α-ScpB antiserum on strains BSG1470 (mparS-amyE) (top panel) and BSG1469 (parS-amyE) (middle panel). Reads were mapped to 5-kb bins. Signals for IP samples were divided by the signals of the normalized input. Ratios were calculated by dividing the values of the parS-amyE strain by the mparS-amyE strain (bottom panel). A number above one indicates more reads in the parS-amyE sample (shown in the blue colors), for all other windows, the inverse ratios were calculated and displayed in gray colors.
See also Figure S6.