Figure 7.
Model for the Recruitment of Smc/ScpAB to and Release from parS Sites
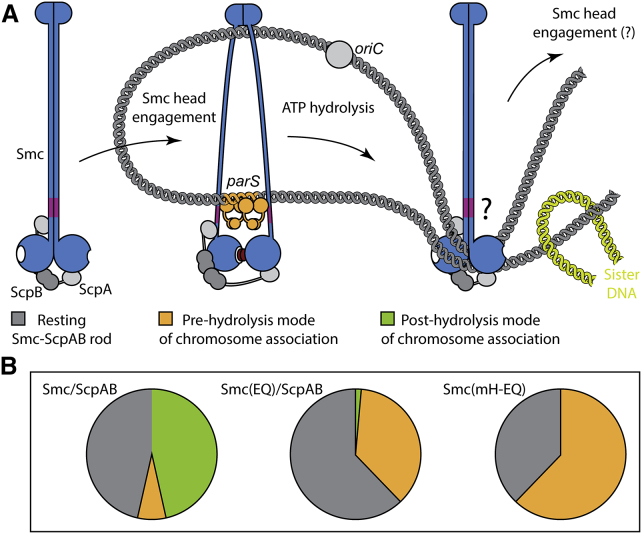
(A) Model for the targeting to and release from ParB/parS by holo-Smc/ScpAB. Most Smc/ScpAB exists as a rod-shaped structure, which is unable to bind to DNA via its hinge or to ParB/parS via the coiled coils. Dissolution of the Smc rod and engagement of Smc head domains are prerequisites for the targeting of Smc/ScpAB to parS. Upon ATP hydrolysis, the ring-like structure might revert to the rod conformation and is released from parS DNA. Sister DNA segments (in green colors) might be excluded from the Smc rod due to steric restrictions. Repetitive rod-ring-rod transitions might drive DNA loop extrusion.
(B) Pie charts displaying rough estimates for the relative occupancy of the different states illustrated in (A) based on Smc head cross-linking efficiency (Figure 4B). Please note that the fraction of wild-type Smc complexes on and off the chromosome (depicted as green and gray pies in the left chart) is unknown. A tiny fraction of chromosomally loaded Smc(EQ)/ScpAB has been detected (Wilhelm et al., 2015).