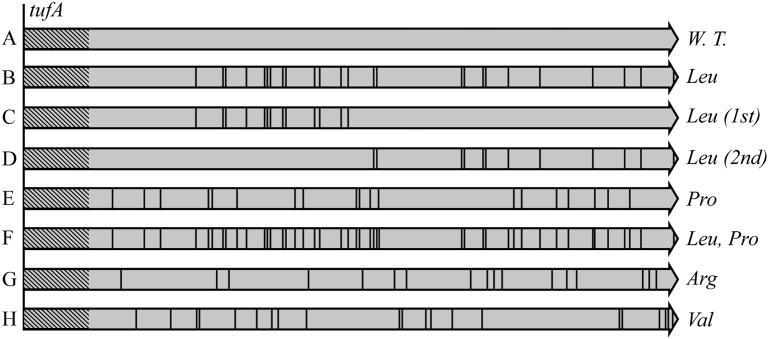
Fig 1. Overview of the design of synonymous tuf alleles.
The first forty codons in tuf were left unchanged to reduce the possible impact of N-terminal codon usage bias (hatched region). Black bars represent codons that have been changed to synonymous codons. (A) Wild-type tufA gene. (B) Leucine codons (N = 25) were changed to UUA, UUG, CUU, CUC or CUA, respectively. (C) The leucine codons in the first half of the tuf gene (N = 13) were changed to UUA, CUC or CUA, respectively. (D) The leucine codons in the second half of the tuf gene (N = 12) were changed to UUA, CUC or CUA, respectively. (E) Proline codons (N = 19) were changed to CCU, CCC or CCA. (F) Both leucine and proline codons (N = 44) were changed: in one tuf allele all leucine codons were changed to UUG and all proline codons to CCU; in the other tuf allele all leucine codons were changed to UUA and all proline codons to CCA. (G) All arginine CGU codons (N = 17) were changed to CGG. (H) All valine GUU codons (N = 21) were changed to GUC.