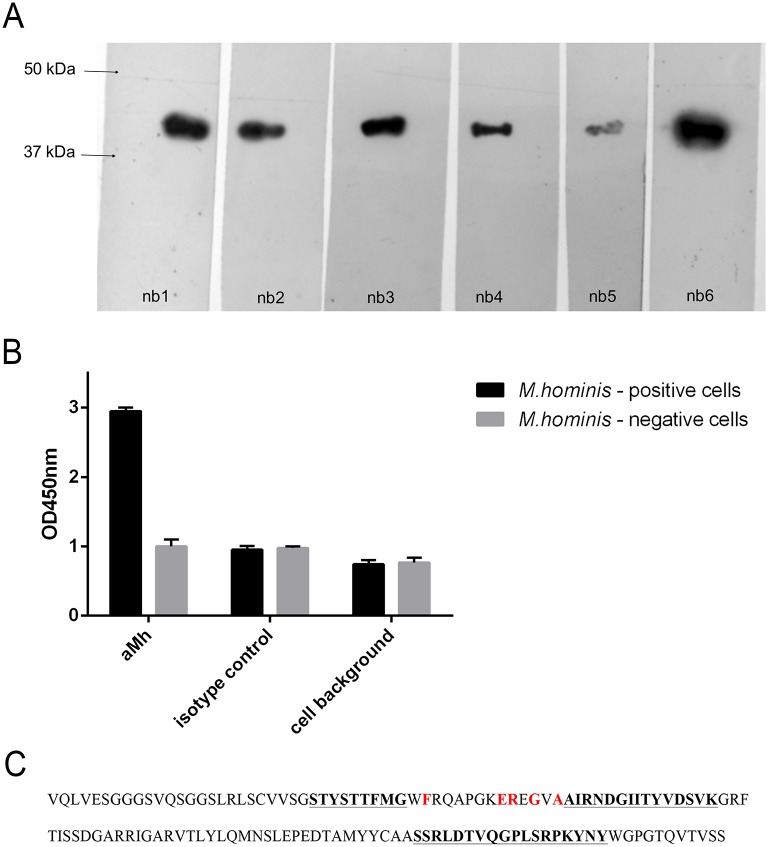
Fig 1.
A—Western blot analysis of binding selected nanobodies to LAMPs from M. hominis. LAMPs from M. hominis H-34 was boiled with Laemmli sample buffer, loaded on 12% Laemmli SDS-PAGE; and Western blot was performed for binding with nanobodies (nb 1–6) at working concentration 1 mkg/ml. B—Cell-based ELISA analysis of binding selected nanobody nb 6 (named aMh) to mycoplasma infected cells. M. hominis positive (experimentally infected) and control M. hominis negative A549 cells were fixed with paraformaldehyde and ELISA with selected aMh (nb6) was performed. aMh—incubation with aMh, isotype control—incubation with control nanobody, cells background—fixed cells background. C—Amino acid sequence of the selected nanobody aMh. Hypervariable regions CDR1, CDR2 and CDR3 are underlined and highlighted in bold. Positions of characteristic amino acids that are specific for single domain antibodies are highlighted in red (these amino acids differ in variable domains of classical type antibodies).