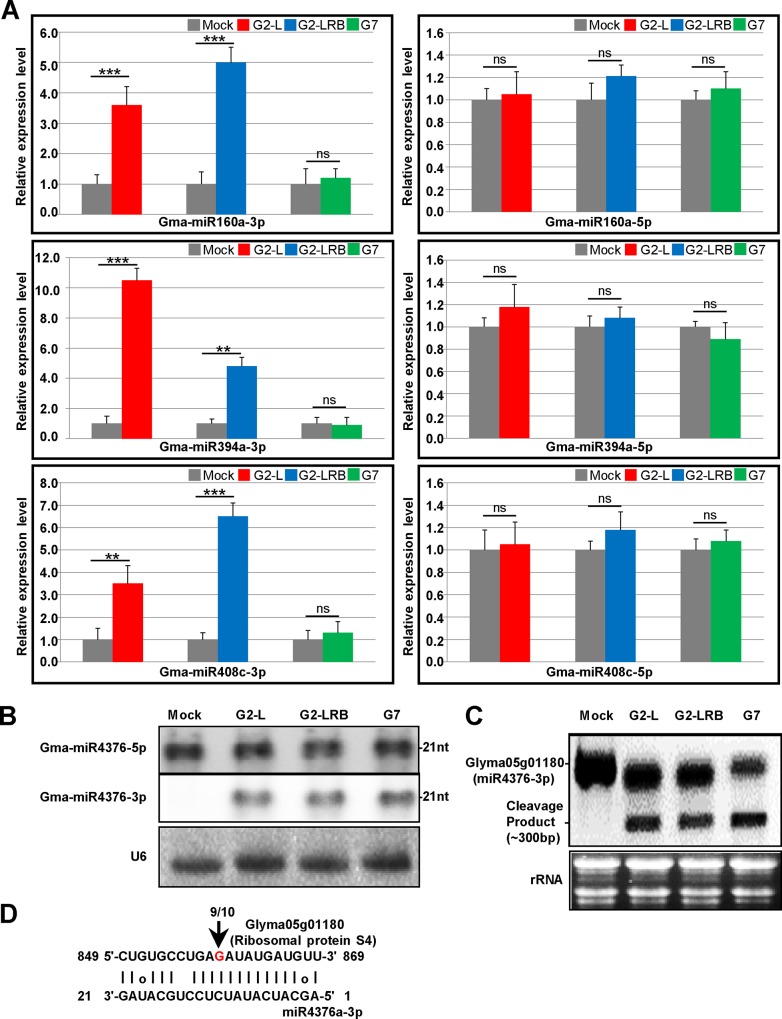
Fig 4. The enhanced accumulation of miRNA-3p in SMV-infected soybean plants.
(A) Expression analysis of miRNA-5p and miRNA-3p in mock-inoculated and SMV-infected soybean plants at 14 dpi by stem-loop RT-qPCR analysis. Soybean 18S rRNA was used as an internal control. Error bars represent mean ± SD (standard deviation) and the data are averages from three biological replicates. Asterisks indicate statistically significant differences comparing with the mock control (student’s t-tests) (*p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant). (B) Expression analysis of miRNA-43763p by Northern blots. Soybean U6 was used as an internal control to normalize miRNA accumulation. (C) Northern blot analysis of the expression of GLYMA05g01180, a predicted target of miR4376-3p. The rRNA stained with ethidium bromide was used as a loading control. (D) Mapping of the cleavage site in GLYMA05g01180 by RLM-5' RACE assay. The numbers above the arrows indicate the frequencies of sequenced RACE clones corresponding to the cleavage site.