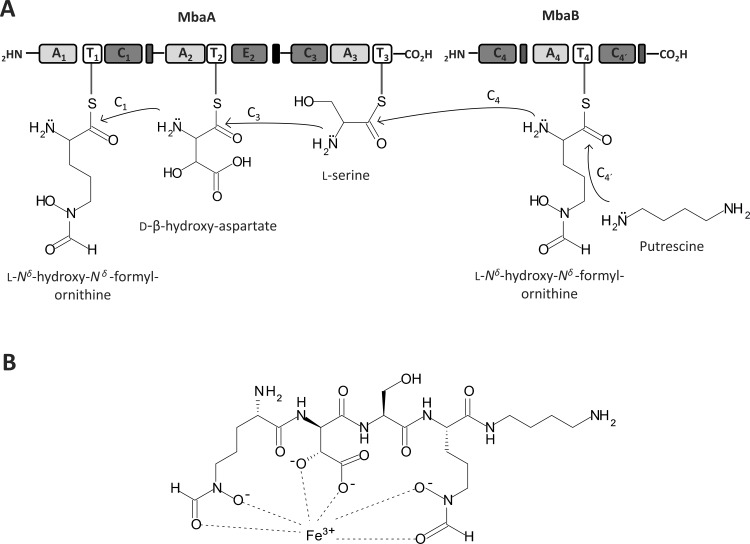
Fig 5. Siderophore synthesis by the non-ribosomal peptide synthetases MbaA and MbaB in B. xenovorans LB400 and the ferrisiderophore complex structure.
A, Modular organization of the non-ribosomal peptide synthetases encoded by the mbaA and mbaB genes. Domains are represented as blocks. Adenylation (A), thiolation (T), condensation (C) and epimerization (E) domains of MbaA are organized in three modules. Domains of MbaB are organized in one module. The synthesis of the siderophore starts with specific substrates assembly to each NRPS module. Substrate of modules 1 and 4 is L-Nδ-hydroxy-Nδ-formylornithine (hfOrn). Substrates of module 2 and 3 are D-β-hydroxy-aspartic acid (OH-Asp) and L-serine (Ser), respectively. Condensation domains of each module allow the attachment of the four substrates to form the tetrapeptide. C1 condenses hfOrn with OH-Asp, C3 condenses OH-Asp with Ser and C4 condenses Ser with hfOrn. Chemical modifications of the amino acids are catalyzed by the protein products of mbaH, mbaC and mbaE genes before the assembly to each NRPS module. The α-ketoglutarate-dependent hydroxylase MbaH hydroxylates the β-carbon of Asp. The L-ornithine-5-monooxygenase encoded by the mbaC gene hydroxylates the nitrogen δ of both Orn. The formyltranserase encoded by the mbaE gene formylates the nitrogen δ of Orn 1 and 4. C4´ domain of module 4 adds a diaminobutane (putrescine) molecule in the N-terminus of the modified tetrapeptide. B, Ferrisiderophore complex structure synthesized by B. xenovorans LB400. The ferrisiderophore structure was predicted by bioinformatic analyses and confirmed by ESI-MS and MALDI-TOF MS of the complex purified by HPLC.