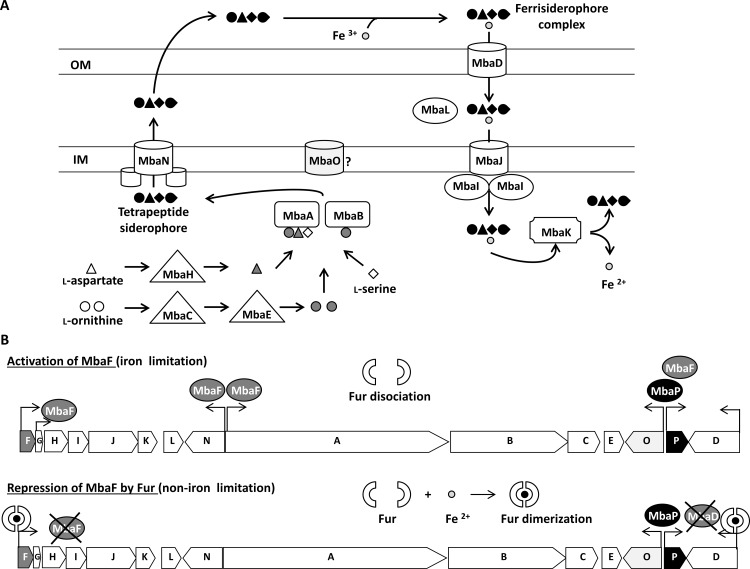
Fig 8. Model for the biosynthesis, transport and regulation of the non-ribosomal peptide siderophore in B. xenovorans LB400 based on bioinformatic analyses.
A, The amino acids are modified by MbaH, MbaC and MbaE enzymes and assembled by MbaA and MbaB NRPS that synthetize the siderophore. The siderophore is exported through MbaN transporter localized in the inner membrane (IM). The siderophore binds iron(III) in the extracellular space, forming the ferrisiderophore complex. The complex is recognized by MbaD in the outer membrane (OM) and imported through the inner membrane by the MbaIJL ABC transporter. From the cytoplasmic ferrisiderophore complex the iron is released by MbaK. MbaO MFS transporter may be involved in siderophore transport, but its function is unknown. B, The transcriptional expression of the mba gene cluster from B. xenovorans LB400 is repressed by iron. In presence of iron, dimerized Fur binds to the Fur box present in the mbaF and mbaD genes and prevents their transcription. In absence of iron, Fur inactivation released ECF sigma factor binding site of mbaF gene, which allows the transcription of the mbaF gene. MbaF factor binds to the MbaF boxes in the mbaG, mbaN, mbaA and mbaP genes promoters for their expression. The mbaP gene transcription is also expressed in iron-rich condition. The LysR-type transcriptional regulator MbaP binds to the LysR binding site present in the mbaP and mbaO genes allowing their expression.